阿里天池赛后数据分析
赛后数据分析
天池精准医疗大赛——人工智能辅助糖尿病遗传风险预测
大赛概况
进入21世纪,生命科学特别是基因科技已经广泛而且深刻影响到每个人的健康生活,于此同时,科学家们借助基因科技史无前例的用一种全新的视角解读生命和探究疾病本质。人工智能(AI)能够处理分析海量医疗健康数据,通过认知分析获取洞察,服务于政府、健康医疗机构、制药企业及患者,实现个性化,可以循证的智慧医疗,推动创新,实现价值。
心血管病、糖尿病等慢性疾病,每年导致的死亡人数占总死亡人数的80%,每年用于慢病医疗费用占中国公共医疗卫生支出的比例超过13%。作为一种常见慢性疾病,糖尿病目前无法根治,但却能通过科学有效的干预、预防和治疗,来降低发病率和提高患者的生活质量。阿里云联合青梧桐健康科技有限公司主办天池精准医疗大赛——人工智能辅助糖尿病遗传风险预测,希望用人工智能的方法和思想处理、分析、解读和应用糖尿病相关大数据,让参赛选手设计高精度,高效,且解释性强的算法来挑战糖尿病精准预测这一科学难题,为学术界和精准医疗提供有力的技术支撑,帮助我们攻克糖尿病。
from pylab import *
mpl.rcParams['font.sans-serif'] = ['Droid Sans Fallback']
mpl.rcParams['axes.unicode_minus'] = Falseimport pandas as pd
data = pd.read_csv(r'data_flie/d_train_20180102.csv')
data.columnsIndex([u'id', u'性别', u'年龄', u'体检日期', u'*天门冬氨酸氨基转换酶', u'*丙氨酸氨基转换酶', u'*碱性磷酸酶',
u'*r-谷氨酰基转换酶', u'*总蛋白', u'白蛋白', u'*球蛋白', u'白球比例', u'甘油三酯', u'总胆固醇',
u'高密度脂蛋白胆固醇', u'低密度脂蛋白胆固醇', u'尿素', u'肌酐', u'尿酸', u'乙肝表面抗原', u'乙肝表面抗体',
u'乙肝e抗原', u'乙肝e抗体', u'乙肝核心抗体', u'白细胞计数', u'红细胞计数', u'血红蛋白', u'红细胞压积',
u'红细胞平均体积', u'红细胞平均血红蛋白量', u'红细胞平均血红蛋白浓度', u'红细胞体积分布宽度', u'血小板计数',
u'血小板平均体积', u'血小板体积分布宽度', u'血小板比积', u'中性粒细胞%', u'淋巴细胞%', u'单核细胞%',
u'嗜酸细胞%', u'嗜碱细胞%', u'血糖'],
dtype='object')
describe_df = data.describe()
describe_df
| id | 年龄 | *天门冬氨酸氨基转换酶 | *丙氨酸氨基转换酶 | *碱性磷酸酶 | *r-谷氨酰基转换酶 | *总蛋白 | 白蛋白 | *球蛋白 | 白球比例 | ... | 血小板计数 | 血小板平均体积 | 血小板体积分布宽度 | 血小板比积 | 中性粒细胞% | 淋巴细胞% | 单核细胞% | 嗜酸细胞% | 嗜碱细胞% | 血糖 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 5642.000000 | 5642.000000 | 4421.000000 | 4421.00000 | 4421.000000 | 4421.000000 | 4421.000000 | 4421.000000 | 4421.000000 | 4421.000000 | ... | 5626.000000 | 5619.000000 | 5619.000000 | 5619.000000 | 5626.000000 | 5626.000000 | 5626.000000 | 5626.000000 | 5626.000000 | 5642.000000 |
| mean | 2866.184155 | 45.580468 | 26.805892 | 27.65202 | 87.482278 | 38.823762 | 76.771108 | 45.822504 | 30.948604 | 1.502538 | ... | 253.085318 | 10.653444 | 13.304414 | 0.267907 | 56.718468 | 33.778546 | 6.856772 | 2.043797 | 0.603697 | 5.631925 |
| std | 1655.555649 | 12.961946 | 13.563003 | 22.61152 | 25.630897 | 40.788282 | 4.016055 | 2.617401 | 3.565997 | 0.220621 | ... | 59.586828 | 0.985484 | 2.173716 | 0.060503 | 7.799940 | 7.253097 | 1.567583 | 1.710901 | 0.291760 | 1.544882 |
| min | 1.000000 | 3.000000 | 10.040000 | 0.12000 | 22.980000 | 6.360000 | 57.320000 | 29.540000 | 7.060000 | 0.520000 | ... | 37.000000 | 7.100000 | 8.000000 | 0.042000 | 14.400000 | 7.500000 | 3.100000 | 0.000000 | 0.000000 | 3.070000 |
| 25% | 1433.250000 | 35.000000 | 20.250000 | 15.13000 | 70.370000 | 17.810000 | 74.190000 | 44.130000 | 28.570000 | 1.360000 | ... | 213.000000 | 10.000000 | 11.700000 | 0.230000 | 51.600000 | 28.800000 | 5.800000 | 0.900000 | 0.400000 | 4.920000 |
| 50% | 2870.500000 | 45.000000 | 23.890000 | 21.48000 | 84.470000 | 26.190000 | 76.630000 | 45.820000 | 30.780000 | 1.490000 | ... | 249.000000 | 10.600000 | 12.900000 | 0.260000 | 56.700000 | 33.600000 | 6.700000 | 1.600000 | 0.600000 | 5.290000 |
| 75% | 4302.750000 | 54.000000 | 29.270000 | 32.30000 | 100.210000 | 43.850000 | 79.530000 | 47.570000 | 33.180000 | 1.630000 | ... | 289.000000 | 11.300000 | 14.600000 | 0.300000 | 62.000000 | 38.500000 | 7.700000 | 2.600000 | 0.700000 | 5.767500 |
| max | 5732.000000 | 93.000000 | 434.950000 | 498.89000 | 374.320000 | 736.990000 | 100.410000 | 54.080000 | 66.180000 | 7.120000 | ... | 745.000000 | 15.200000 | 25.300000 | 0.710000 | 88.500000 | 76.300000 | 23.200000 | 22.500000 | 3.500000 | 38.430000 |
8 rows × 40 columns
data.isnull().sum() / len(data)#缺失值比例id 0.000000
性别 0.000000
年龄 0.000000
体检日期 0.000000
*天门冬氨酸氨基转换酶 0.216413
*丙氨酸氨基转换酶 0.216413
*碱性磷酸酶 0.216413
*r-谷氨酰基转换酶 0.216413
*总蛋白 0.216413
白蛋白 0.216413
*球蛋白 0.216413
白球比例 0.216413
甘油三酯 0.216058
总胆固醇 0.216058
高密度脂蛋白胆固醇 0.216058
低密度脂蛋白胆固醇 0.216058
尿素 0.244240
肌酐 0.244240
尿酸 0.244240
乙肝表面抗原 0.758419
乙肝表面抗体 0.758419
乙肝e抗原 0.758419
乙肝e抗体 0.758419
乙肝核心抗体 0.758419
白细胞计数 0.002836
红细胞计数 0.002836
血红蛋白 0.002836
红细胞压积 0.002836
红细胞平均体积 0.002836
红细胞平均血红蛋白量 0.002836
红细胞平均血红蛋白浓度 0.002836
红细胞体积分布宽度 0.002836
血小板计数 0.002836
血小板平均体积 0.004077
血小板体积分布宽度 0.004077
血小板比积 0.004077
中性粒细胞% 0.002836
淋巴细胞% 0.002836
单核细胞% 0.002836
嗜酸细胞% 0.002836
嗜碱细胞% 0.002836
血糖 0.000000
dtype: float64可以看出与乙肝相关的特征,缺失值到达了75%以上,于是决定删除乙肝5项,id与血糖无关,也删除,然后将data分为特征和结果标签,并将空缺值用平均值代替,之前打算剔除了体检日期这一特征,想了想等等吧
import time
import datetime
from dateutil.parser import parse
data['体检日期'] = (pd.to_datetime(data['体检日期']) - parse('2017-10-09')).dt.days
data['性别'] = data['性别'].map({'男': 1, '女': 0})
train_lable = data['血糖']#提取标签
train_data = data.iloc[:, 0:-1]#提取特征
exclude_other = ['id', '乙肝表面抗原', '乙肝表面抗体', '乙肝e抗原', '乙肝e抗体', '乙肝核心抗体']
for i in exclude_other:
del train_data[i]
data_mean = train_data.mean()
train_data = train_data.fillna(data_mean)
train_data.shape(5642, 35)import matplotlib.pylab as plt
import seaborn as sns
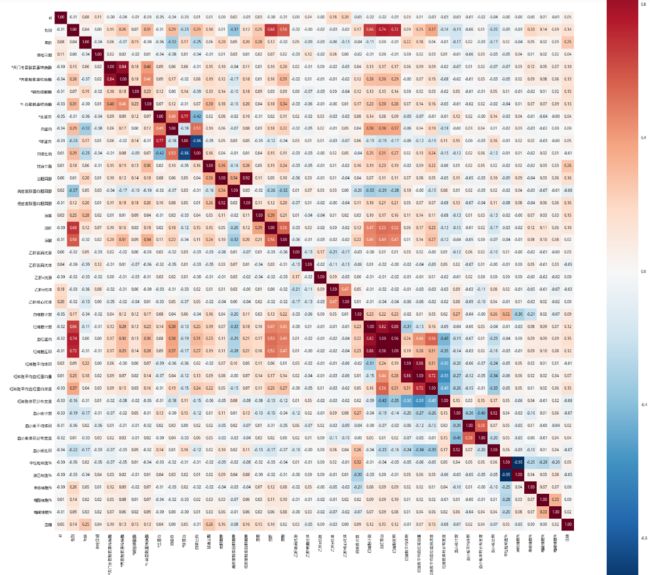
data_corr = data.corr()
fig = plt.figure(figsize=(30, 30))
sns.heatmap(data_corr, vmax=0.9, square=True, cbar=True, annot=True, fmt='.2f', annot_kws={'size': 10})
plt.show()
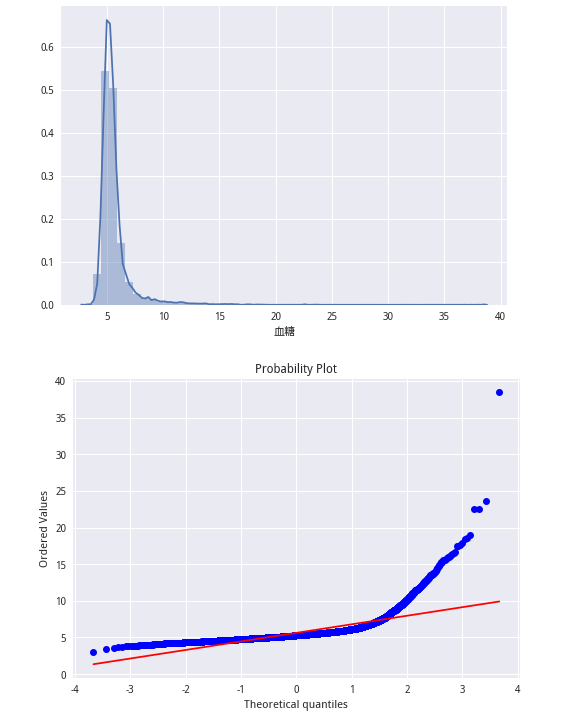
from scipy import stats
sns.distplot(train_lable)
plt.show()
stats.probplot(train_lable, plot=plt)
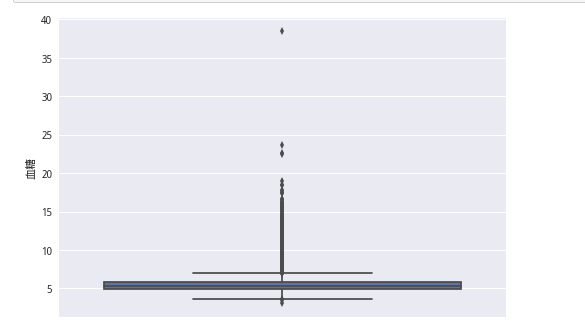
plt.show()sns.boxplot(y = train_lable)
plt.show()剔除血糖超過20的
del_index = []
for i in range(train_lable.shape[0]):
if train_lable[i] > 20:
del_index.append(i)
train_lable.drop(train_lable.index[del_index], inplace=True)
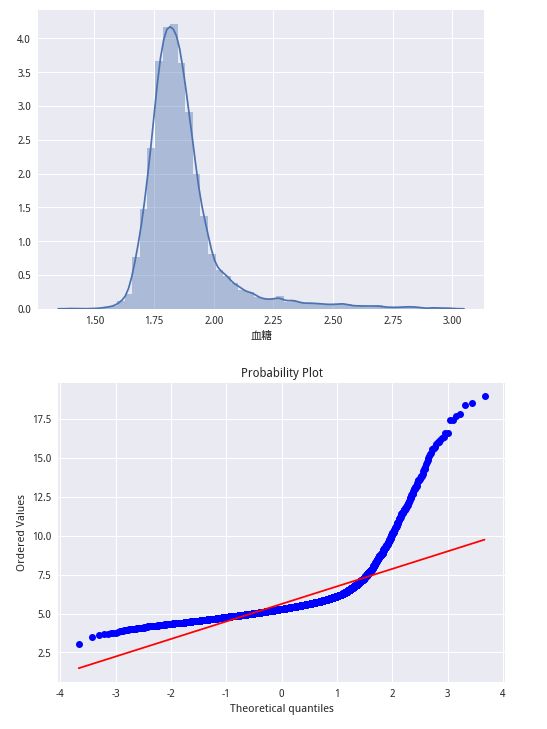
train_data.drop(train_data.index[del_index], inplace=True)import numpy as np
train_lable_log = np.log1p(train_lable)
sns.distplot(train_lable_log);
plt.show()
stats.probplot(train_lable, plot=plt)
plt.show()
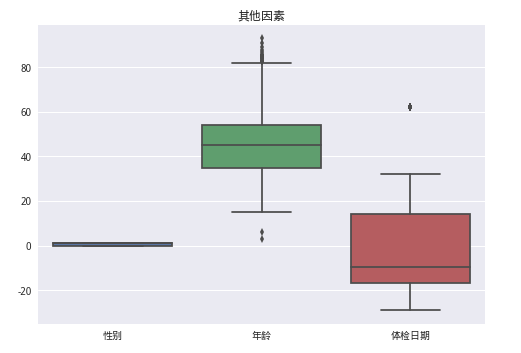
data1 = train_data[['性别', '年龄', '体检日期']]
sns.boxplot(data1)
plt.title('其他因素')
plt.show()data2 = train_data[['*天门冬氨酸氨基转换酶', '*丙氨酸氨基转换酶', '*碱性磷酸酶',
'*r-谷氨酰基转换酶', '*总蛋白', '白蛋白', '*球蛋白']]
sns.boxplot(data2)
plt.title('酶蛋白')
plt.show()train_data.drop(train_data[(train_data['*天门冬氨酸氨基转换酶'] > 200) | (train_data['*丙氨酸氨基转换酶'] > 250) |
(train_data['*r-谷氨酰基转换酶'] > 600)].index, inplace=True)data3 = train_data[['甘油三酯', '总胆固醇','高密度脂蛋白胆固醇', '低密度脂蛋白胆固醇']]
sns.boxplot(data3)
plt.title('醇')
plt.show()
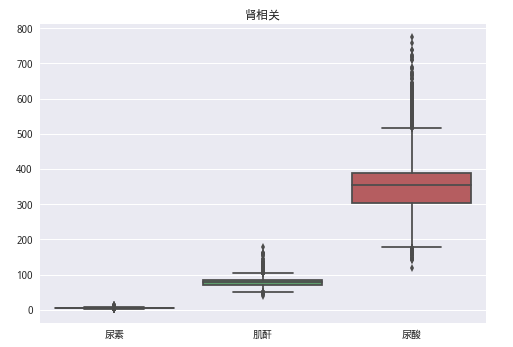
train_data.drop(train_data[(train_data['甘油三酯'] > 30) | (train_data['总胆固醇'] > 20)].index, inplace=True)data4 = train_data[['尿素', '肌酐', '尿酸']]
sns.boxplot(data4)
plt.title('肾相关')
plt.show()
data5 = train_data[['白细胞计数', '红细胞计数', '血红蛋白', '红细胞压积', '红细胞平均体积', '红细胞平均血红蛋白量', '红细胞平均血红蛋白浓度',
'红细胞体积分布宽度', '血小板计数']]
sns.boxplot(data5)
plt.title('細胞數')
plt.show()
train_data.drop(train_data[(train_data['血小板计数'] > 600)].index, inplace=True)data6 = train_data[['血小板平均体积', '血小板体积分布宽度', '血小板比积']]
sns.boxplot(data6)
plt.title('血小板')
plt.show()
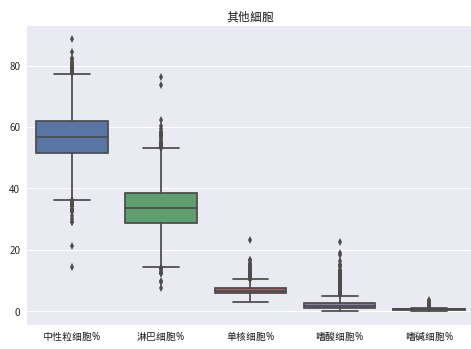
data7 = train_data[['中性粒细胞%','淋巴细胞%', '单核细胞%', '嗜酸细胞%', '嗜碱细胞%']]
sns.boxplot(data7)
plt.title('其他細胞')
plt.show()
train_data.drop(train_data[(train_data['中性粒细胞%'] < 23) | (train_data['淋巴细胞%'] > 65) | (train_data['单核细胞%'] >20)].index, inplace=True)drop_col = ['嗜碱细胞%','单核细胞%','白球比例','白蛋白','*总蛋白', '低密度脂蛋白胆固醇', '血小板比积','淋巴细胞%']
train_data.drop((drop_col), axis=1, inplace=True)fig = plt.figure(figsize=(20, 20))
data_corr = train_data.corr()
sns.heatmap(data_corr, vmax=0.9, square=True)
plt.show()
train_data = train_data.fillna(-999)def data_yunsun(train, test):
train['霉'] = train['*天门冬氨酸氨基转换酶'] + train['*丙氨酸氨基转换酶'] + train['*碱性磷酸酶'] + train['*r-谷氨酰基转换酶']
test['霉'] = test['*天门冬氨酸氨基转换酶'] + test['*丙氨酸氨基转换酶'] + test['*碱性磷酸酶'] + test['*r-谷氨酰基转换酶']
train['尿酸/肌酐'] = train['尿酸'] / train['肌酐']
test['尿酸/肌酐'] = test['尿酸'] / test['肌酐']
train['肾'] = train['尿酸'] + train['尿素'] + train['肌酐']
test['肾'] = test['尿酸'] + test['尿素'] + test['肌酐']
train['红细胞计数*红细胞平均血红蛋白量'] = train['红细胞计数'] * train['红细胞平均血红蛋白量']
test['红细胞计数*红细胞平均血红蛋白量'] = test['红细胞计数'] * test['红细胞平均血红蛋白量']
train['红细胞计数*红细胞平均血红蛋白浓度'] = train['红细胞计数'] * train['红细胞平均血红蛋白浓度']
test['红细胞计数*红细胞平均血红蛋白浓度'] = test['红细胞计数'] * test['红细胞平均血红蛋白浓度']
train['红细胞计数*红细胞平均体积'] = train['红细胞计数'] * train['红细胞平均体积']
test['红细胞计数*红细胞平均体积'] = test['红细胞计数'] * test['红细胞平均体积']
train['嗜酸细胞'] = train['白细胞计数'] * train['嗜酸细胞%']
test['嗜酸细胞'] = test['白细胞计数'] * test['嗜酸细胞%']
train['血红蛋白/红细胞计数*红细胞平均血红蛋白浓度'] = train['血红蛋白'] / train['红细胞计数*红细胞平均血红蛋白浓度']
test['血红蛋白/红细胞计数*红细胞平均血红蛋白浓度'] = test['血红蛋白'] / test['红细胞计数*红细胞平均血红蛋白浓度']
return train, test
'''catboost'''
def test_ans(X_train, Y_train, X_test):
cat_feature_inds = []
cat_feature_inds.append(0)
num_ensembles = 5
y_pred = 0.0
for i in tqdm(range(num_ensembles)):
model = CatBoostRegressor(
iterations=1000, learning_rate=0.03,
depth=6, l2_leaf_reg=3,
loss_function='RMSE',
eval_metric='RMSE',
random_seed=i)
model.fit(X_train, Y_train,cat_features=[0])
y_pred += model.predict(X_test)
y_pred /= num_ensembles
submission = pd.DataFrame({'pred': y_pred})
submission.to_csv(r'sub{}.csv'.format(datetime.datetime.now().strftime('%Y%m%d_%H%M%S')), header=None,
index=False, float_format='%.4f')'''xgboost'''
def test_ans2(X_train, y_train):
import xgboost as xgb
from sklearn.metrics import mean_squared_error
kf = KFold(n_splits=5, shuffle=False)
params = {
'max_depth': 6,
'eta': 0.1,
'silent': 1,
'gamma':0.0468,
'alpha':0.4640,
'lambda': 0.8571,
'objective': 'count:poisson'
}
print "*********"
y_pred = 0.0
for train_index, test_index in kf.split(X_train):
dtrain = xgb.DMatrix(X_train[train_index], y_train[train_index])
dtest = xgb.DMatrix(X_train[test_index], y_train[test_index])
dtest2 = xgb.DMatrix(X_test)
watch_list = [(dtest, 'eval'), (dtrain, 'train')]
num_rounds = 1000
model = xgb.train(params, dtrain, num_rounds, watch_list)
ans = model.predict(dtest)
# score += ((ans-y_train[test_index])**2).sum()/(2*y_train[test_index].shape[0])
y_pred += model.predict(dtest2, ntree_limit=model.best_ntree_limit)
print "+++++++++++"
y_pred /= 5
submission = pd.DataFrame({'pred': y_pred})
submission.to_csv(r'sub{}.csv'.format(datetime.datetime.now().strftime('%Y%m%d_%H%M%S')), header=None,
index=False, float_format='%.4f')
'''ligthGBM'''
def test_ans3(X_train, y_train, X_test):
import lightgbm as lgb
kf = KFold(n_splits=5, shuffle=False)
params = {
'learning_rate': 0.01,
'boosting_type': 'gbdt',
'objective': 'poisson',
'bagging_fraction': 0.8,
'bagging_freq':1,
'num_leaves': 12,
'colsample_bytree': 0.6,
'max_depth': 6,
'min_data': 5,
'min_hessian': 1,
'verbose': -1
}
score = 0.0
ans = 0.0
for train_index, test_index in kf.split(X_train):
lgb_train = lgb.Dataset(X_train[train_index], y_train[train_index])
lgb_eval = lgb.Dataset(X_train[test_index], y_train[test_index], reference=lgb_train)
# lgb_test = lgb.Dataset(X_test)
gbm = lgb.train(params,
lgb_train,
num_boost_round=20000,
valid_sets=lgb_eval,
verbose_eval=500,
early_stopping_rounds=200)
y_pred = gbm.predict(X_train[test_index], num_iteration=gbm.best_iteration)
ans += gbm.predict(X_test, num_iteration=gbm.best_iteration)
score += ((y_pred-y_train[test_index])**2).sum()/(2*y_train[test_index].shape[0])
print score/5
ans /= 5
submission = pd.DataFrame({'pred': ans})
submission.to_csv(r'sub{}.csv'.format(datetime.datetime.now().strftime('%Y%m%d_%H%M%S')), header=None,
index=False, float_format='%.4f')