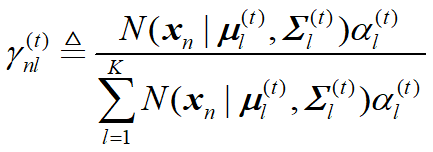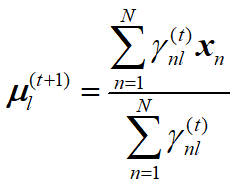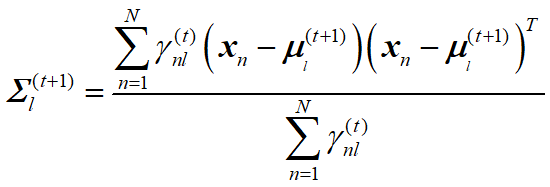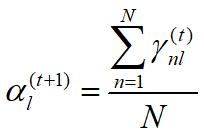EM算法估计GMM (二维高斯混合模型)参数 python 2.7(西瓜书版)
#!/usr/bin/python
# coding:utf-8
# 19-5-23 上午10:07
# @File : EM.py
import numpy as np
import math
import matplotlib.pyplot as plt
#real parm
p = [0.2, 0.3, 0.5] #选择高斯分布的概率
mean1 = [1.0, 2.0]
mean2 = [2.0, 1.0]
mean3 = [1.0, 1.0]
cov1 = [[1.0, 0], [0, 10.0]]
cov2 = [[10.0, 0], [0, 1.0]]
cov3 = [[3.0, 0], [0, 4.0]]
# estimate parm
Emean1 = np.array([1.0, 1.0])
Emean2 = np.array([1.0, 1.0])
Emean3 = np.array([1.0, 1.0])
Ecov1 = np.array([[1.0, 0.0], [0.0, 10.0]])
Ecov2 = np.array([[2.0, 0.0], [0.0, 3.0]])
Ecov3 = np.array([[3.0, 0.0], [0.0, 4.0]])
Ep = p = [0.2, 0.3, 0.5]
# saver parameter
# E 步保存的第l 个模型的 R(n,l)
parameter_dict={}
# 分别存放第l 个模型的 R(n,l)
parameter_dict["gama_list1"] = []
parameter_dict["gama_list2"] = []
parameter_dict["gama_list3"] = []
def DenityOfNorm(Data, Emean,Ecov):
# return EM估计参数 下 data (x, y)处 的正态函数密度
# Data 输入的数据的点
#Emean ,EM算法估计的参数
#Ecov ,EM在第t步估计的协方差矩阵
#
# print Ecov.shape
#
Ecov_sqrt = math.sqrt(np.linalg.det(Ecov)) #协方差矩阵行列式值的开平方
Ecov_inv = np.linalg.inv(Ecov) #协方差矩阵的逆
#we
Data =np.reshape(Data,[2,1])
Emean =np.reshape(Emean,[2,1])
Minus_Data = Data - Emean
Minus_Data_T = Minus_Data.T
res = (1.0 / (2.0 * math.pi * Ecov_sqrt)) * math.exp(
(-0.5) * (np.dot(np.dot(Minus_Data_T, Ecov_inv), Minus_Data)))
return res
# sigma_sqrt = math.sqrt(np.linalg.det(sigma)) # 协方差矩阵绝对值的1/2次
# sigma_inv = np.l
# 生成二维高斯模型的数据
def ini_Normal(mean, cov):
# 返回一个由size指定形状的数组,数组中的值服从
# μ = loc, σ = scale
data =np.random.multivariate_normal(mean, cov,1)
return data
#生成高斯混合模型的数据
def ini_data(k=1,parm1=0.2,parm2=0.3):
data= []
for i in range(k):
d = np.random.rand(1)
if d < parm1:
data.append(ini_Normal(mean1, cov1))
elif d < parm1+parm2:
data.append(ini_Normal(mean2, cov2))
else:
data.append(ini_Normal(mean3, cov3))
return data
#第E步
def E(Data):
# !!!!!!!!!!!!!!!!
# 这里一定要清空, 不然会很尴尬, 之前第三个忘了清空,以为陷入局部最优值,检查了很久
# !!!!!!!!!!!!!!!
global parameter_dict
parameter_dict["gama_list1"] = []
parameter_dict["gama_list2"] = []
parameter_dict["gama_list3"] = []
for point in Data:
# gama_i = (pw * PDF(point, mu_2, sigma_2)) / (
# (1.0 - pw) * PDF(point, mu_1, sigma_1) + pw * PDF(point, mu_2, sigma_2))
# parameter_dict["gama_list"].append(gama_i)
gam_sum = Ep[0] * DenityOfNorm(point,Emean1,Ecov1)+Ep[1] * DenityOfNorm(point,Emean2,Ecov2)+Ep[2] * DenityOfNorm(point,Emean3,Ecov3)
gam1_i = Ep[0] * DenityOfNorm(point,Emean1,Ecov1) / gam_sum
parameter_dict["gama_list1"].append(gam1_i)
gam2_i = Ep[1] * DenityOfNorm(point,Emean2, Ecov2) / gam_sum
parameter_dict["gama_list2"].append(gam2_i)
parameter_dict["gama_list3"].append(1.0-gam2_i-gam1_i)
#第M步
def M(Data):
r1 = 0
r2 = 0
r3 = 0
for i in range(len(parameter_dict['gama_list2'])):
r1 = r1 + parameter_dict["gama_list1"][i]
r2 = r2 + parameter_dict["gama_list2"][i]
r3 = r3 + parameter_dict["gama_list3"][i]
# r3 += 1- parameter_dict["gama_list1"][i]- parameter_dict["gama_list2"][i]
# updata estimated p
# for i in range(3):
Ep[0] = r1 / len(Data)
Ep[1] = r2 / len(Data)
Ep[2] = r3 / len(Data)
# updata estimated mean
Emean1_new= np.array([0.0, 0.0])
Emean2_new= np.array([0.0, 0.0])
Emean3_new= np.array([0.0, 0.0])
#
Emean1_new.shape = (1,2)
Emean2_new.shape = (1,2)
Emean3_new.shape = (1,2)
# print parameter_dict["gama_list1"].shape
for i in range(len(Data)):
print i
# print parameter_dict["gama_list1"][i]
Emean1_new =Emean1_new + Data[i] * parameter_dict["gama_list1"][i] /r1
Emean2_new =Emean2_new + Data[i] * parameter_dict["gama_list2"][i] /r2
Emean3_new =Emean3_new + Data[i] * parameter_dict["gama_list3"][i] /r3
# Emean3_new += Data[i] * (1-parameter_dict["gama_list1"][i]-parameter_dict["gama_list2"])
global Emean1
global Emean2
global Emean3
# print "---------------------------------------"
# print Emean1_new.shape
Emean1 = Emean1_new
Emean2 = Emean2_new
Emean3 = Emean3_new
# updata estimated cov
Ecov1_new = np.array([[0, 0], [0, 0]])
Ecov2_new = np.array([[0, 0], [0, 0]])
Ecov3_new = np.array([[0, 0], [0, 0]])
for i in range(len(Data)):
Data_MiuMean1 = Data[i] - Emean1
Data_MiuMean2 = Data[i] - Emean2
Data_MiuMean3 = Data[i] - Emean3
Data_MiuMean1_T = np.array(Data_MiuMean1).T
Data_MiuMean2_T = np.array(Data_MiuMean2).T
Data_MiuMean3_T = np.array(Data_MiuMean3).T
# 测试
# MYtry = np.dot(Data_MiuMean2,Data_MiuMean2_T)
# MYtry1 = np.dot(Data_MiuMean2_T,Data_MiuMean2)
# print MYtry, MYtry.shape
# print MYtry1, MYtry1.shape
#
Ecov1_new = Ecov1_new + np.dot(Data_MiuMean1_T, Data_MiuMean1) * parameter_dict["gama_list1"][i]
Ecov2_new = Ecov2_new + np.dot(Data_MiuMean2_T, Data_MiuMean2) * parameter_dict["gama_list2"][i]
Ecov3_new = Ecov3_new + np.dot(Data_MiuMean3_T, Data_MiuMean3) * parameter_dict["gama_list3"][i]
# Ecov2_new = Ecov2_new + np.d
global Ecov1
global Ecov2
global Ecov3
# print "------------------------"
# print Ecov1_new.shape
Ecov1 = Ecov1_new / r1
Ecov2 = Ecov2_new / r2
Ecov3 = Ecov3_new / r3
#不可以控制迭代次数EM算法
def EM_iterate( Data, esp=0.0001):
# Data:数据
#param esp:终止约束,第n次优化和第n+1次优化差距小于0.0001
# set_parameter(mu_1, sigma_1, mu_2, sigma_2, pi_weight)
while (True):
old_m1 = Emean1
old_m2 = Emean2
old_m3 = Emean3
E(Data)
M(Data)
#
# print Emean2.shape
#
delta_1 = Emean1 - old_m1
delta_2 = Emean2 - old_m2
delta_3 = Emean3 - old_m3
# print delta_1 ,delta_1.shape
# delta_2 = E - old_m2
if math.fabs(delta_1[0][0]) < esp and math.fabs(delta_1[0][1]) < esp and math.fabs(
delta_2[0][0]) < esp and math.fabs(delta_2[0][1]) < esp and math.fabs(
delta_3[0][0]) < esp and math.fabs(delta_3[0][1]) < esp:
break
#EM 算法可以控制迭代次数
def EM_iterat_N(iterm, Data,esp=0.0001):
# 记录更新的过程
mean1_trace = [[], []]
mean2_trace = [[], []]
mean3_trace = [[], []]
# mean2_trace = [[], []]
# mean3_trace = [[], []]
for i in range(iterm):
old_m1 = Emean1
old_m2 = Emean2
old_m3 = Emean3
E(Data)
M(Data)
#
# print Emean2.shape
#
mean1_trace[0].append(Emean1[0][0])
mean1_trace[1].append(Emean1[0][1])
mean2_trace[0].append(Emean2[0][0])
mean2_trace[1].append(Emean2[0][1])
mean3_trace[0].append(Emean3[0][0])
mean3_trace[1].append(Emean3[0][1])
delta_1 = Emean1 - old_m1
delta_2 = Emean2 - old_m2
delta_3 = Emean3 - old_m3
# print delta_1 ,delta_1.shape
# delta_2 = E - old_m2
if math.fabs(delta_1[0][0]) < esp and math.fabs(delta_1[0][1]) < esp and math.fabs(delta_2[0][0]) < esp and math.fabs(delta_2[0][1]) < esp and math.fabs(delta_3[0][0]) < esp and math.fabs(delta_3[0][1]) < esp:
break
# print mean1_trace
# plt.subplot(121)
# plt.xlim(xmax=5, xmin=2)
# plt.ylim(ymax=90, ymin=60)
# plt.xlabel("eruptions")
# plt.ylabel("waiting")
# plt.plot(mean1_trace[:,0], mean1_trace[:,1], 'r-')
# plt.plot(mean1_trace[:,0],mean1_trace[:,1], 'b^')
#
# plt.subplot(122)
# plt.xlim(xmax=4, xmin=0)
# plt.ylim(ymax=60, ymin=40)
# plt.xlabel("eruptions")
# plt.ylabel("waiting")
# plt.plot(mean2_trace[:,0], mean2_trace[:,1], 'r-')
# plt.plot(mean2_trace[:,0], mean2_trace[:,1], 'bo')
# plt.show()
return mean1_trace,mean2_trace,mean3_trace
def main():
# data = gen_clusters()
# save_data(data, '3clusters.txt')
# d = load_data('3clusters.txt')
# show_scatter(d)
# # show_scatter(d)
data = ini_data(10000)
#print data
Emean1.shape = (1,2)
Emean2.shape = (1,2)
Emean3.shape = (1,2)
# EM_iterate( data)
trace1,trace2,trace3 = EM_iterat_N(50, data)
# 画图
plt.subplot(131)
plt.xlim(xmax=5, xmin=2)
plt.ylim(ymax=90, ymin=60)
plt.xlabel("Emean1_x")
plt.ylabel("Emean1_y")
plt.plot(trace1[0], trace1[1], 'r-')
plt.plot(trace1[0], trace1[1], 'b^')
plt.axis('tight')
plt.subplot(132)
plt.xlim(xmax=4, xmin=0)
plt.ylim(ymax=60, ymin=40)
plt.xlabel("Emean2_x")
plt.ylabel("Emean2_y")
plt.plot(trace2[0], trace2[1], 'r-')
plt.plot(trace2[0], trace2[1], 'bo')
plt.axis('tight')
plt.subplot(133)
plt.xlim(xmax=4, xmin=0)
plt.ylim(ymax=60, ymin=40)
plt.xlabel("Emean3_x")
plt.ylabel("Emean3_y")
plt.plot(trace3[0], trace3[1], 'r-')
plt.plot(trace3[0], trace3[1], 'bv')
plt.axis('tight')
plt.show()
print "real mean1: ", mean1
print "real mean1: ", mean2
print "real mean1: ", mean3
print "estimate means:", Emean1, Emean2, Emean3
if __name__ == '__main__':
main()
更新公式: