【SA-SSD】点云数据和gt_bbox可视化
文章目录
- 一. 使用mayavi原因
- 二. 读取bin文件显示
- 1. 代码
- 2. 结果
- 三. SA-SSD中点云和gt_bboxs显示
- 1. 准备工作
- 2. 代码
- 2. 结果
一. 使用mayavi原因
由于torch和open3d冲突,使用torch的时候open3d就使用不了,但是我还是想看一下可视化的点云数据,于是就用mayavi进行可视化。如果有知道怎么解决open3d和torch冲突的朋友,也请留言~
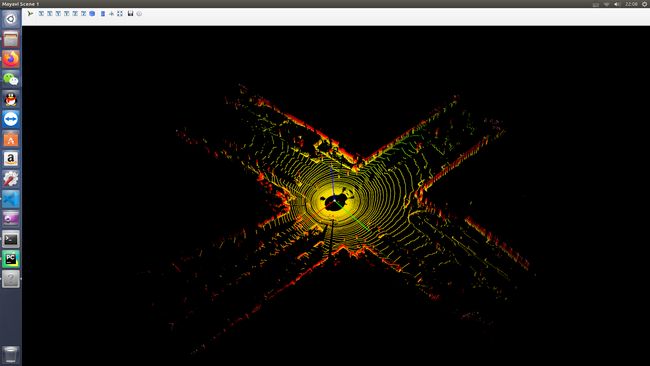
二. 读取bin文件显示
1. 代码
import numpy as np
import mayavi.mlab
#mayavi显示点云
def read_pointcloud_from_bin_file(bin_path):
pointcloud = np.fromfile(bin_path, dtype=np.float32).reshape(-1,4)
print(pointcloud.shape)
print(type(pointcloud))
x = pointcloud[:, 0] # x position of point
xmin = np.amin(x, axis=0)
xmax = np.amax(x, axis=0)
y = pointcloud[:, 1] # y position of point
ymin = np.amin(y, axis=0)
ymax = np.amax(y, axis=0)
z = pointcloud[:, 2] # z position of point
zmin = np.amin(z, axis=0)
zmax = np.amax(z, axis=0)
print(xmin,xmax,ymin,ymax,zmin,zmax)
d = np.sqrt(x ** 2 + y ** 2) # Map Distance from sensor
vals = 'height'
if vals == "height":
col = z
else:
col = d
fig = mayavi.mlab.figure(bgcolor=(0, 0, 0), size=(640, 500))
mayavi.mlab.points3d(x, y, z,
col, # Values used for Color
mode="point",
# 灰度图的伪彩映射
colormap='spectral', # 'bone', 'copper', 'gnuplot'
# color=(0, 1, 0), # Used a fixed (r,g,b) instead
figure=fig,
)
# 绘制原点
mayavi.mlab.points3d(0, 0, 0, color=(1, 1, 1), mode="sphere",scale_factor=1)
# 绘制坐标
axes = np.array(
[[20.0, 0.0, 0.0, 0.0], [0.0, 20.0, 0.0, 0.0], [0.0, 0.0, 20.0, 0.0]],
dtype=np.float64,
)
#x轴
mayavi.mlab.plot3d(
[0, axes[0, 0]],
[0, axes[0, 1]],
[0, axes[0, 2]],
color=(1, 0, 0),
tube_radius=None,
figure=fig,
)
#y轴
mayavi.mlab.plot3d(
[0, axes[1, 0]],
[0, axes[1, 1]],
[0, axes[1, 2]],
color=(0, 1, 0),
tube_radius=None,
figure=fig,
)
#z轴
mayavi.mlab.plot3d(
[0, axes[2, 0]],
[0, axes[2, 1]],
[0, axes[2, 2]],
color=(0, 0, 1),
tube_radius=None,
figure=fig,
)
mayavi.mlab.show()
def plot3Dbox(corners):
for i in range(corners.shape[0]):
corner = corners[i]
idx = np.array([0, 1, 2, 3, 0, 4, 5, 6, 7, 4, 5, 1, 2, 6, 7, 3])
x = corner[0, idx]
y = corner[1, idx]
z = corner[2, idx]
mayavi.mlab.plot3d(x, y, z, color=(0.23, 0.6, 1), colormap='Spectral', representation='wireframe', line_width=5)
# mlab.show(stop=True)
def main():
corners = np.array([[[0.0, 1, 1, 0, 0, 1, 1, 0], [0, 0, 1, 1, 0, 0, 1, 1], [0, 0, 0, 0, 1, 1, 1, 1]]])
plot3Dbox(corners)
read_pointcloud_from_bin_file("/home/seivl/data/KITTI/training/velodyne/004965.bin")
if __name__ == '__main__':
main()
2. 结果
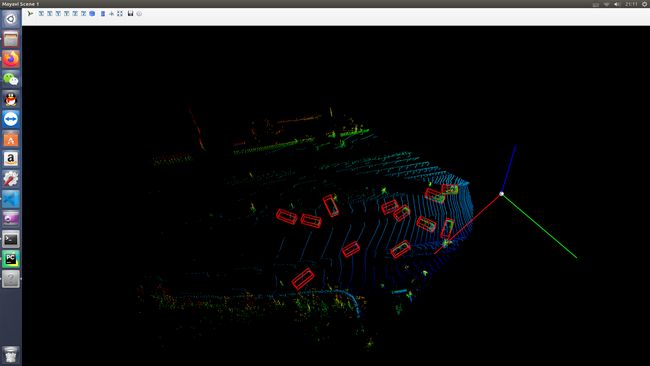
三. SA-SSD中点云和gt_bboxs显示
1. 准备工作
在 kitti.py 这个模块的 def prepare_train_img() 函数最后,加入下面一段代码:
if True:
test_data = {}
test_data['points'] = points
test_data['gt_bboxes'] = gt_bboxes
test_data['gt_types'] = gt_types
import pickle
with open("/home/seivl/test_data.pkl", 'wb') as fo:
pickle.dump(test_data, fo)
2. 代码
import numpy as np
import mayavi.mlab
import pickle
def corners_nd(dims, origin=0.5):
"""generate relative box corners based on length per dim and
origin point.
Args:
dims (float array, shape=[N, ndim]): array of length per dim
origin (list or array or float): origin point relate to smallest point.
Returns:
float array, shape=[N, 2 ** ndim, ndim]: returned corners.
point layout example: (2d) x0y0, x0y1, x1y0, x1y1;
(3d) x0y0z0, x0y0z1, x0y1z0, x0y1z1, x1y0z0, x1y0z1, x1y1z0, x1y1z1
where x0 < x1, y0 < y1, z0 < z1
"""
ndim = int(dims.shape[1])
corners_norm = np.stack(
np.unravel_index(np.arange(2 ** ndim), [2] * ndim), axis=1).astype(
dims.dtype)
# now corners_norm has format: (2d) x0y0, x0y1, x1y0, x1y1
# (3d) x0y0z0, x0y0z1, x0y1z0, x0y1z1, x1y0z0, x1y0z1, x1y1z0, x1y1z1
# so need to convert to a format which is convenient to do other computing.
# for 2d boxes, format is clockwise start with minimum point
# for 3d boxes, please draw lines by your hand.
if ndim == 2:
# generate clockwise box corners
corners_norm = corners_norm[[0, 1, 3, 2]]
elif ndim == 3:
corners_norm = corners_norm[[0, 1, 3, 2, 4, 5, 7, 6]]
corners_norm = corners_norm - np.array(origin, dtype=dims.dtype)
corners = dims.reshape([-1, 1, ndim]) * corners_norm.reshape(
[1, 2 ** ndim, ndim])
return corners
def rotation_3d_in_axis(points, angles, axis=0):
# points: [N, point_size, 3]
rot_sin = np.sin(angles)
rot_cos = np.cos(angles)
ones = np.ones_like(rot_cos)
zeros = np.zeros_like(rot_cos)
if axis == 1:
rot_mat_T = np.stack([[rot_cos, zeros, -rot_sin], [zeros, ones, zeros],
[rot_sin, zeros, rot_cos]])
elif axis == 2 or axis == -1:
rot_mat_T = np.stack([[rot_cos, -rot_sin, zeros],
[rot_sin, rot_cos, zeros], [zeros, zeros, ones]])
elif axis == 0:
rot_mat_T = np.stack([[zeros, rot_cos, -rot_sin],
[zeros, rot_sin, rot_cos], [ones, zeros, zeros]])
else:
raise ValueError("axis should in range")
return np.einsum('aij,jka->aik', points, rot_mat_T)
def center_to_corner_box3d(centers, origin=[0.5, 0.5, 0], axis=2):
"""convert kitti locations, dimensions and angles to corners
Args:
centers (float array, shape=[N, 3]): locations in kitti label file.
dims (float array, shape=[N, 3]): dimensions in kitti label file.
angles (float array, shape=[N]): rotation_y in kitti label file.
origin (list or array or float): origin point relate to smallest point.
use [0.5, 1.0, 0.5] in camera and [0.5, 0.5, 0] in lidar.
axis (int): rotation axis. 1 for camera and 2 for lidar.
Returns:
[type]: [description]
"""
# 'length' in kitti format is in x axis.
# yzx(hwl)(kitti label file)<->xyz(lhw)(camera)<->z(-x)(-y)(wlh)(lidar)
# center in kitti format is [0.5, 1.0, 0.5] in xyz.
corners = corners_nd(centers[:,3:6], origin=origin)
corners = rotation_3d_in_axis(corners, centers[:,-1], axis=axis)
corners += centers[:,:3].reshape([-1, 1, 3])
return corners
def plot3Dbox(corners):
for i in range(corners.shape[0]):
corner = corners[i]
idx = np.array([0, 1, 2, 3, 0, 4, 5, 6, 7, 4, 5, 1, 2, 6, 7, 3])
x = corner[0, idx]
y = corner[1, idx]
z = corner[2, idx]
# mayavi.mlab.plot3d(x, y, z, color=(0.23, 0.6, 1), colormap='Spectral', representation='wireframe', line_width=5)
mayavi.mlab.plot3d(x, y, z, color=(1, 0, 0), colormap='Spectral', representation='wireframe')
mayavi.mlab.show()
def show_points(gt_data):
pointcloud = gt_data['points']
print(pointcloud.shape)
print(type(pointcloud))
x = pointcloud[:, 0] # x position of point
xmin = np.amin(x, axis=0)
xmax = np.amax(x, axis=0)
y = pointcloud[:, 1] # y position of point
ymin = np.amin(y, axis=0)
ymax = np.amax(y, axis=0)
z = pointcloud[:, 2] # z position of point
zmin = np.amin(z, axis=0)
zmax = np.amax(z, axis=0)
print(xmin,xmax,ymin,ymax,zmin,zmax)
d = np.sqrt(x ** 2 + y ** 2) # Map Distance from sensor
vals = 'height'
if vals == "height":
col = z
else:
col = d
fig = mayavi.mlab.figure(bgcolor=(0, 0, 0), size=(640, 500))
mayavi.mlab.points3d(x, y, z,
col, # Values used for Color
mode="point",
# 灰度图的伪彩映射
colormap='spectral', # 'bone', 'copper', 'gnuplot'
# color=(0, 1, 0), # Used a fixed (r,g,b) instead
figure=fig,
)
# 绘制原点
mayavi.mlab.points3d(0, 0, 0, color=(1, 1, 1), mode="sphere",scale_factor=1)
# 绘制坐标
axes = np.array(
[[20.0, 0.0, 0.0, 0.0], [0.0, 20.0, 0.0, 0.0], [0.0, 0.0, 20.0, 0.0]],
dtype=np.float64,
)
#x轴
mayavi.mlab.plot3d(
[0, axes[0, 0]],
[0, axes[0, 1]],
[0, axes[0, 2]],
color=(1, 0, 0),
tube_radius=None,
figure=fig,
)
#y轴
mayavi.mlab.plot3d(
[0, axes[1, 0]],
[0, axes[1, 1]],
[0, axes[1, 2]],
color=(0, 1, 0),
tube_radius=None,
figure=fig,
)
#z轴
mayavi.mlab.plot3d(
[0, axes[2, 0]],
[0, axes[2, 1]],
[0, axes[2, 2]],
color=(0, 0, 1),
tube_radius=None,
figure=fig,
)
corners = center_to_corner_box3d(gt_data['gt_bboxes'])
corners = np.array(corners).transpose(0, 2, 1)
plot3Dbox(np.array(corners))
mayavi.mlab.show()
def main():
with open("/home/seivl/test_data.pkl", 'rb') as fo:
gt_data = pickle.load(fo)
show_points(gt_data)
if __name__ == '__main__':
main()