菜鸟入门_Python_机器学习(4)_PCA和MDA降维和聚类
@sprt
*写在开头:博主在开始学习机器学习和Python之前从未有过任何编程经验,这个系列写在学习这个领域一个月之后,完全从一个入门级菜鸟的角度记录我的学习历程,代码未经优化,仅供参考。有错误之处欢迎大家指正。
系统:win7-CPU;
编程环境:Anaconda2-Python2.7,IDE:pycharm5;
参考书籍:
《Neural Networks and Learning Machines(Third Edition)》- Simon Haykin;
《Machine Learning in Action》- Peter Harrington;
《Building Machine Learning Systems with Python》- Wili Richert;
C站里都有资源,也有中文译本。
我很庆幸能跟随老师从最基础的东西学起,进入机器学习的世界。*
降维和聚类算是无监督学习的重要领域,还是那句话,不论是PCA、MDA还是K-means聚类,网上大牛总结的杠杠的,给几个参考链接:
http://www.cnblogs.com/jerrylead/archive/2011/04/18/2020209.html
http://bbezxcy.iteye.com/blog/2090591
http://www.tuicool.com/articles/7nIvum
http://www.cnblogs.com/python27/p/MachineLearningWeek08.html
http://blog.pluskid.org/?p=407
http://www.cnblogs.com/Key-Ky/archive/2013/11/24/3440684.html
http://www.cnblogs.com/coser/archive/2013/04/10/3013044.html
PCA和MDA的推导过程都是手推,本来想拍照发上来,但前几次‘作’过之后实在提不起兴趣,还好有小伙伴(妹子)总结的很好:
http://blog.csdn.net/totodum/article/details/51049165
http://blog.csdn.net/totodum/article/details/51097329
来看我们这次课的任务:
•数据Cat4D3Groups是4维观察数据,
•请先采用MDS方法降维到3D,形成Cat3D3Groups数据,显示并观察。
•对Cat3D3Groups数据采用线性PCA方法降维到2D,形成Cat2D3Groups数据,显示并观察。
•对Cat2D3Groups数据采用K-Mean方法对数据进行分类并最终确定K,显示分类结果。
•对Cat2D3Groups数据采用Hierarchical分类法对数据进行分类,并显示分类结果。
理论一旦推导完成,代码写起来就很轻松:
Part 1:降维处理
MDA:
# -*- coding:gb2312 -*-
from pylab import *
import numpy as np
from mpl_toolkits.mplot3d import Axes3D
def print_D(data):
N = np.shape(data)[0]
d = np.zeros((N, N))
for i in range(N):
c = data[i, :]
for j in range(N):
e = data[j, :]
d[i, j] = np.sqrt(np.sum(np.power(c - e, 2)))
return d
def MDS(D, K):
N = np.shape(D)[0]
D2 = D ** 2
H = np.eye(N) - 1.0/N
T = -0.5 * np.dot(np.dot(H, D2), H)
eigVal, eigVec = np.linalg.eig(T)
indices = np.argsort(eigVal) # 返回从小到大的索引值
indices = indices[::-1] # 反转
eigVal = eigVal[indices] # 特征值从大到小排列
eigVec = eigVec[:, indices] # 排列对应特征向量
m = eigVec[:, :K]
n = np.diag(np.sqrt(eigVal[:K]))
X = np.dot(m, n)
return X
# test
'''
data = genfromtxt("CAT4D3GROUPS.txt")
D = print_D(data)
# print D
# 4D 转 3D
CAT3D3GROUPS = MDS(D, 3)
# print CAT3D3GROUPS
# D_3D = print_D(CAT3D3GROUPS)
# print D_3D
figure(1)
ax = subplot(111,projection='3d')
ax.scatter(CAT3D3GROUPS[:, 0], CAT3D3GROUPS[:, 1], CAT3D3GROUPS[:, 2], c = 'b')
ax.set_zlabel('Z') #坐标轴
ax.set_ylabel('Y')
ax.set_xlabel('X')
title('MDS_4to3')
# 4D 转 2D
CAT2D3GROUPS = MDS(D, 2)
# print CAT2D3GROUPS
# D_2D = print_D(CAT2D3GROUPS)
# print D_2D
figure(2)
plot(CAT2D3GROUPS[:, 0], CAT2D3GROUPS[:, 1], 'b.')
xlabel('x')
ylabel('y')
title('MDS_4to2')
'''
PDA:
# -*- coding:gb2312 -*-
from pylab import *
from numpy import *
from mpl_toolkits.mplot3d import Axes3D
def PCA(data, K):
# 数据标准化
m = mean(data, axis=0) # 每列均值
data -= m
# 协方差矩阵
C = cov(transpose(data))
# 计算特征值特征向量,按降序排序
evals, evecs = linalg.eig(C)
indices = argsort(evals) # 返回从小到大的索引值
indices = indices[::-1] # 反转
evals = evals[indices] # 特征值从大到小排列
evecs = evecs[:, indices] # 排列对应特征向量
evecs_K_max = evecs[:, :K] # 取最大的前K个特征值对应的特征向量
# 产生新的数据矩阵
finaldata = dot(data, evecs_K_max)
return finaldata
# test
'''
data = genfromtxt("CAT4D3GROUPS.txt")
# 4D 转 3D
data_PCA = PCA(data, 3)
# print data_PCA
figure(1)
ax = subplot(111, projection='3d')
ax.scatter(data_PCA[:, 0], data_PCA[:, 1], data_PCA[:, 2], c='b')
ax.set_zlabel('Z') #坐标轴
ax.set_ylabel('Y')
ax.set_xlabel('X')
title('PCA_4to3')
# 4D 转 2D
data_PCA = PCA(data, 2)
print data_PCA
figure(2)
plot(data_PCA[:, 0], data_PCA[:, 1], 'b.')
xlabel('x')
ylabel('y')
title('PCA_4to2')
'''
代码里的注释啰啰嗦嗦应该解释的很清楚,这里不再赘述,看结果:
1、用MDS方法降维到3D,形成Cat3D3Groups数据:
共两个函数,辅助函数用来生成欧氏距离矩阵,MDS函数用于降维。

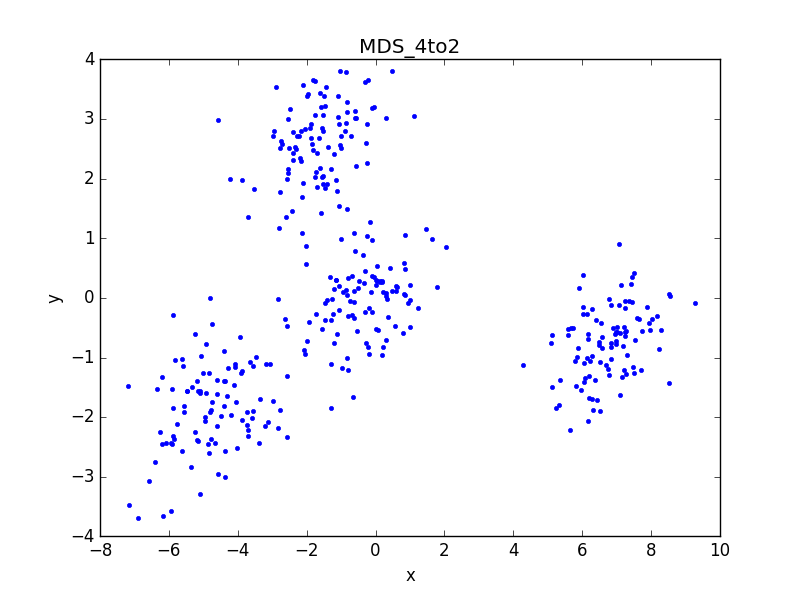
通过输出的距离矩阵可以看出,降维前后欧氏距离误差小于10^-4,证明算法有效。同时旋转3D图像也可以明显找出2D平面图的视角
2、用PCA方法降维到2D,形成Cat2D3Groups数据:


用PCA直接对4D数据降维后的结果与MDS等价,证明算法有效。同时旋转3D图像也可以明显找出2D平面图的视角。
3、总结分析:
先用MDS算法将4D数据降到3D,再用PCA降到2D。

与MDS降维生成的2D图像及数据对比,误差忽略不计,证明算法有效,同时证明MDS和PCA算法在进行小批量数据降维处理上效果类似。
Part 2:聚类分析:
数据用前面降维之后的二维数据。K-means聚类分析的程序主要参考《Machine Learning in Action》- Peter Harrington这本书第十章,我自己添加了选择最优K值的功能:
# -*- coding:gb2312 -*-
import numpy as np
from pylab import *
from numpy import *
# 求欧氏距离
def euclDistance(vector1, vector2):
return np.sqrt(np.sum(np.power(vector2 - vector1, 2)))
# 返回某个值在列表中的全部索引值
def myfind(x, y):
return [ a for a in range(len(y)) if y[a] == x]
# 初始化聚类点
def initCentroids(data, k):
numSamples, dim = data.shape
centroids = np.zeros((k, dim))
for i in range(k):
index = int(np.random.uniform(0, numSamples))
centroids[i, :] = data[index, :]
return centroids
# K-mean 聚类
def K_mean(data, k):
## step 1: 初始化聚点
centroides = initCentroids(data, k)
numSamples = data.shape[0]
clusterAssment = np.zeros((numSamples, 2)) # 保存每个样本点的簇索引值和误差
clusterChanged = True
while clusterChanged:
clusterChanged = False
global sum
sum = []
# 对每一个样本点
for i in xrange(numSamples):
minDist = np.inf # 记录最近距离
minIndex = 0 # 记录聚点
## step 2: 找到距离最近的聚点
for j in range(k):
distance = euclDistance(centroides[j, :], data[i, :])
if distance < minDist:
minDist = distance
minIndex = j
## step 3: 将该样本归到该簇
if clusterAssment[i, 0] != minIndex:
clusterChanged = True # 前后分类相同时停止循环
clusterAssment[i, :] = minIndex, minDist ** 2 # 记录簇索引值和误差
## step 4: 更新聚点
for j in range(k):
index = myfind(j, clusterAssment[:, 0])
pointsInCluster = data[index, :] # 返回属于j簇的data中非零样本的目录值,并取出样本
centroides[j, :] = np.mean(pointsInCluster, axis=0) # 求列平均
# 返回cost funktion值
suml = 0
lenght = pointsInCluster.shape[0]
for l in range(lenght):
dis = euclDistance(centroides[j, :], pointsInCluster[l, :])
suml += dis ** 2 / lenght
sum.append(suml)
cost = np.sum(sum) / k
print cost
return centroides, clusterAssment, cost
# 画出分类前后结果
def showCluster(data, k, centroides, clusterAssment):
numSamples, dim = data.shape
mark = ['r.', 'b.', 'g.', 'k.', '^r', '+r', 'sr', 'dr', ', 'pr']
# draw all samples
for i in xrange(numSamples):
markIndex = int(clusterAssment[i, 0])
figure(2)
plt.plot(data[i, 0], data[i, 1], mark[markIndex])
plt.title('K-means')
mark = ['Dr', 'Db', 'Dg', 'Dk', '^b', '+b', 'sb', 'db', ', 'pb']
# draw the centroids
for i in range(k):
figure(2)
plt.plot(centroides[i, 0], centroides[i, 1], mark[i], markersize = 12)
其中三个辅助函数用于求欧氏距离,返回矩阵索引值和画图,k-mean函数用于聚类,当所有样本点到其所属聚类中心距离不变时,输出聚类结果,并返回cost function的值。
Cost function计算方法:对每个簇,求所有点到所属聚类中心的欧氏距离,平方后取均值E。聚类结束后,所有簇E值求和取平均得到cost function的值。
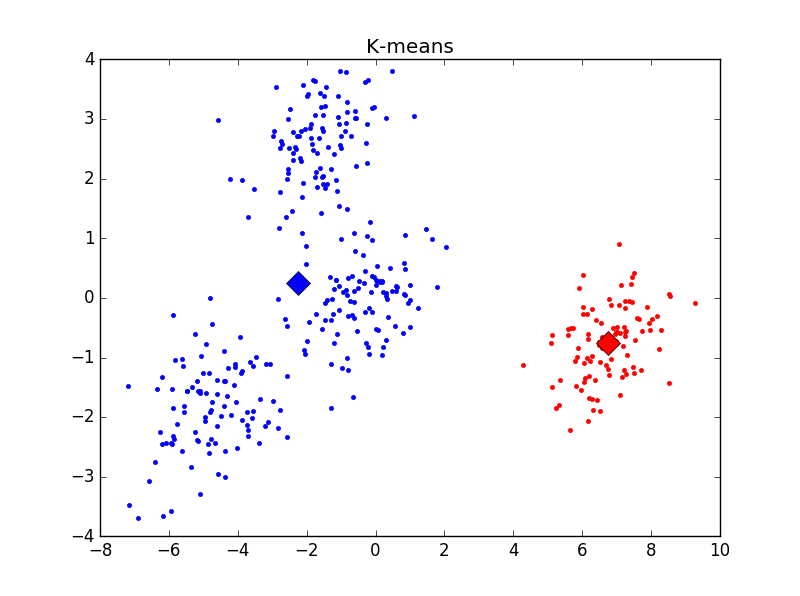
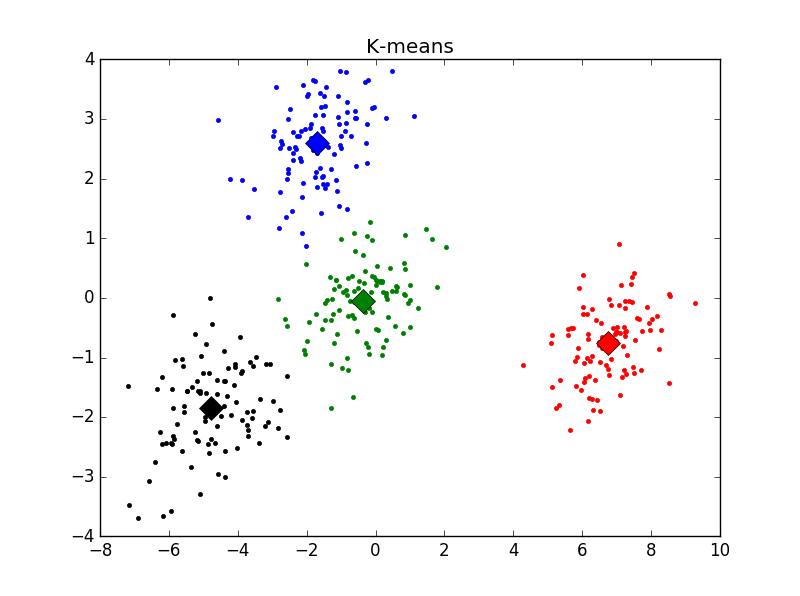
主观判断,k = 4时聚类结果最优。用Elbow方法选择K值结果如下:
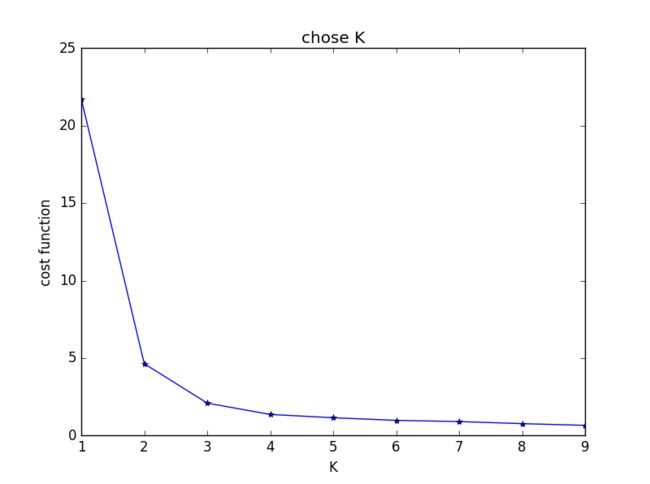
发现在K = 2时cost function值下降最为明显,与之前判断的结果不符。思考后发现,K=1时聚类没有意义,所以上图并不能有效选择K值,调整后结果如下:
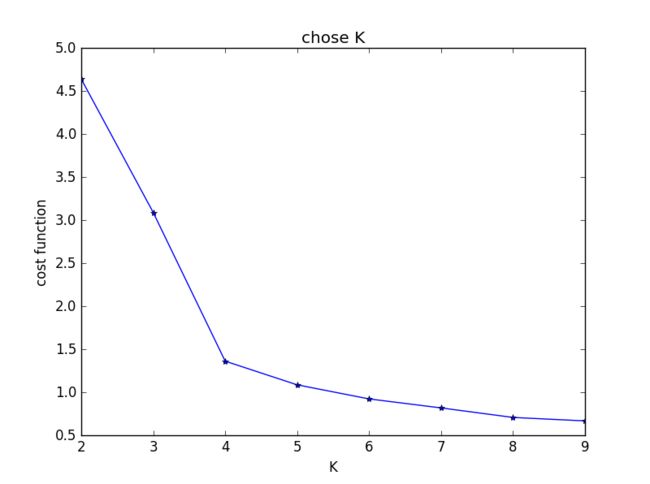
明显看出,k = 4时cost function下降极为明显。与主观判断结果相符。
Hierarchical分类,参考网上代码,出处不记得了:
# -*- coding:gb2312 -*-
import numpy as np
import matplotlib.pyplot as plt
import MDS
import PCA
def yezi(clust):
if clust.left == None and clust.right == None :
return [clust.id]
return yezi(clust.left) + yezi(clust.right)
def Euclidean_distance(vector1,vector2):
length = len(vector1)
TSum = sum([pow((vector1[i] - vector2[i]),2) for i in range(len(vector1))])
SSum = np.sqrt(TSum)
return SSum
def loadDataSet(fileName):
a = []
with open(fileName, 'r') as f:
data = f.readlines() #txt中所有字符串读入data
for line in data:
odom = line.split() #将单个数据分隔开存好
numbers_float = map(float, odom) #转化为浮点数
a.append(numbers_float) #print numbers_float
a = np.array(a)
return a
class bicluster:
def __init__(self, vec, left=None,right=None,distance=0.0,id=None):
self.left = left
self.right = right #每次聚类都是一对数据,left保存其中一个数据,right保存另一个
self.vec = vec #保存两个数据聚类后形成新的中心
self.id = id
self.distance = distance
def list_array(wd, clo):
len_=len(wd)
xc=np.zeros([len_, clo])
for i in range(len_):
ad = wd[i]
xc[i, :] = ad
return xc
def hcluster(data, n) :
[row,column] = data.shape
data = list_array(data, column)
biclusters = [bicluster(vec = data[i], id = i) for i in range(len(data))]
distances = {}
flag = None
currentclusted = -1
while(len(biclusters) > n) : #假设聚成n个类
min_val = np.inf #Python的无穷大
biclusters_len = len(biclusters)
for i in range(biclusters_len-1) :
for j in range(i + 1, biclusters_len):
#print biclusters[i].vec
if distances.get((biclusters[i].id,biclusters[j].id)) == None:
#print biclusters[i].vec
distances[(biclusters[i].id,biclusters[j].id)] = Euclidean_distance(biclusters[i].vec,biclusters[j].vec)
d = distances[(biclusters[i].id,biclusters[j].id)]
if d < min_val:
min_val = d
flag = (i,j)
bic1,bic2 = flag #解包bic1 = i , bic2 = j
newvec = [(biclusters[bic1].vec[i] + biclusters[bic2].vec[i])/2 for i in range(len(biclusters[bic1].vec))] #形成新的类中心,平均
newbic = bicluster(newvec, left=biclusters[bic1], right=biclusters[bic2], distance=min_val, id = currentclusted) #二合一
currentclusted -= 1
del biclusters[bic2] #删除聚成一起的两个数据,由于这两个数据要聚成一起
del biclusters[bic1]
biclusters.append(newbic)#补回新聚类中心
clusters = [yezi(biclusters[i]) for i in range(len(biclusters))] #深度优先搜索叶子节点,用于输出显示
return biclusters,clusters
def showCluster(dataSet, k, num_mark):
numSamples, dim = dataSet.shape
mark = ['r.', 'b.', 'g.', 'k.', '^r', '+r', 'sr', 'dr', ', 'pr']
# draw all samples
for i in xrange(numSamples):
plt.plot(dataSet[i, 0], dataSet[i, 1], mark[num_mark])
plt.xlabel('X')
plt.ylabel('Y')
plt.title('Hierarchical')
if __name__ == "__main__":
# 加载数据
dataMat =np.genfromtxt('CAT4D3GROUPS.txt') #400*4
dataSet = PCA.PCA(dataMat, 2)
k,l = hcluster(dataSet, 10) # l返回了聚类的索引
# 选取规模最大的k个簇,其他簇归为噪音点
for j in range(len(l)):
m = []
for ii in range(len(l[j])):
m.append(l[j][ii])
m = np.array(m)
a = dataSet[m]
showCluster(a,len(l),j)
plt.show()
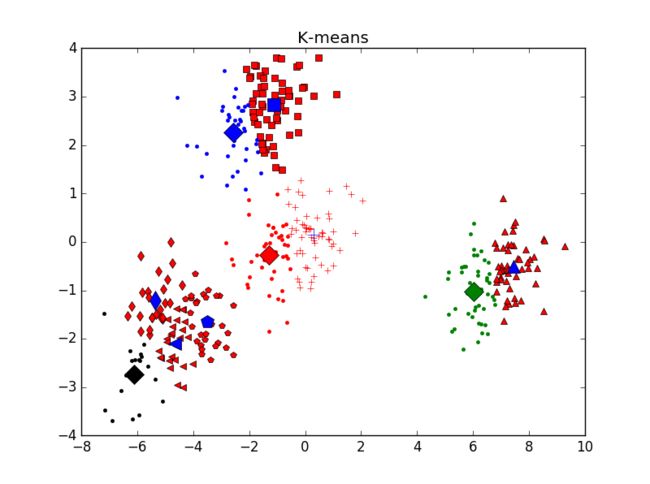
当一个类集合中包含多个样本点时,类与类之间的距离取Group Average:把两个集合中的点两两的欧氏距离全部放在一起求平均值,分类结果如下:
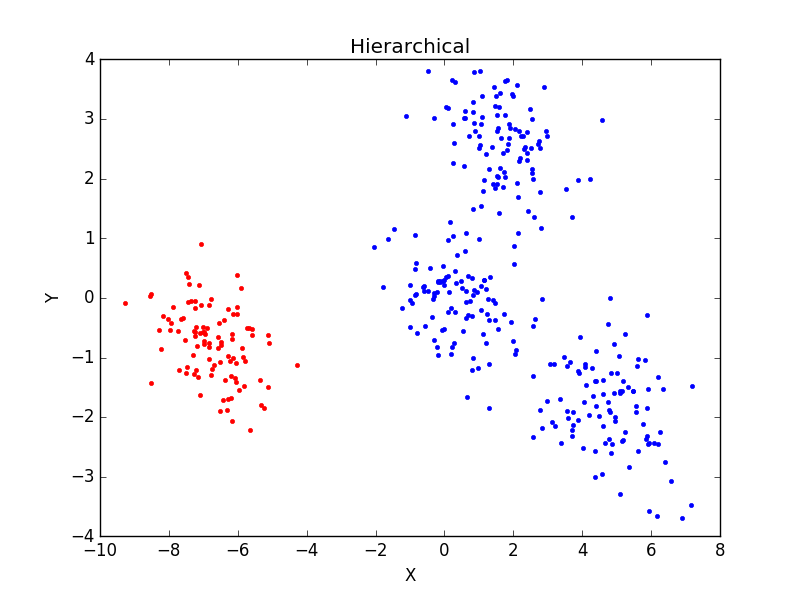
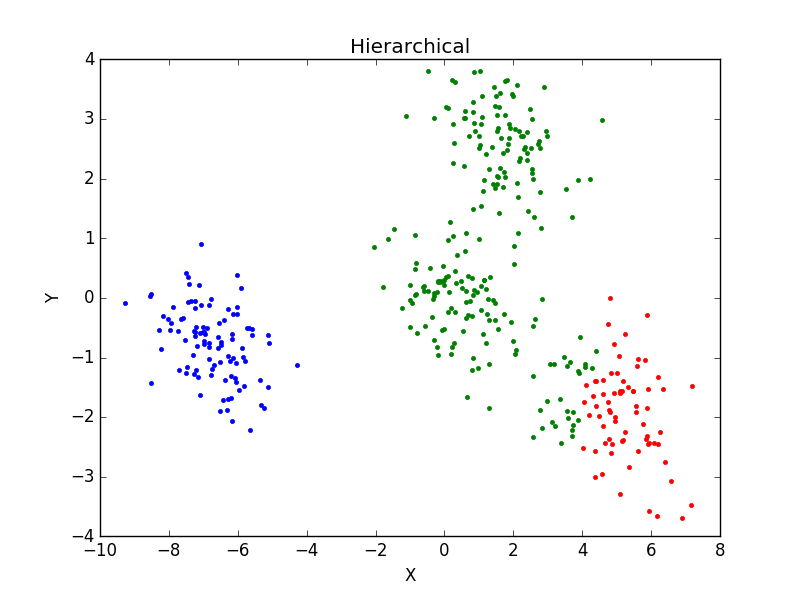

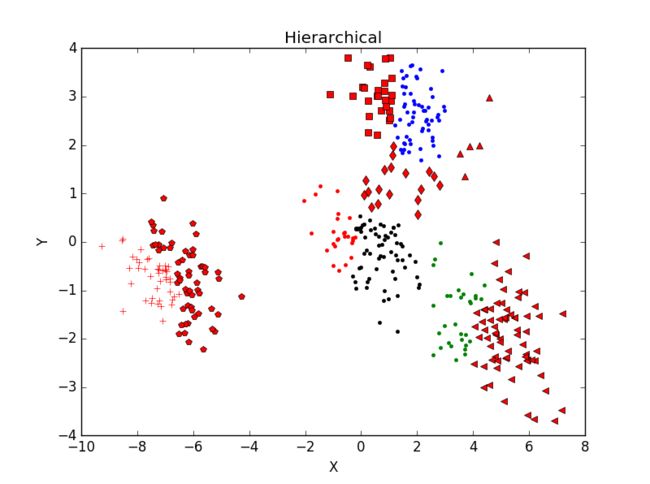
重复运行后分类结果并未有太大变化。主观判断,从分成3类及4类的结果看,Hierarchical分类方法效果不如K-mean聚类效果好。