利用基于线性假设的线性分类器LogisticRegression/SGDClassifier进行二类分类(复习1)
本文是个人学习笔记,内容主要涉及LR(LogisticRegression)和SGD(SGDClassifier)对breast-cancer数据集进行线性二分类。
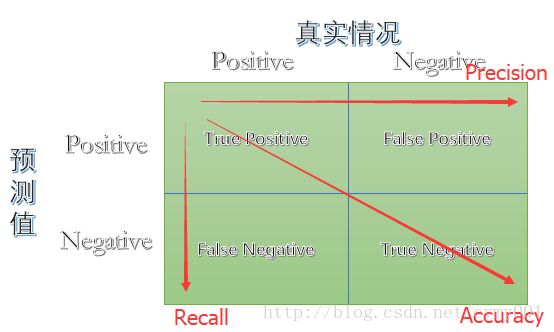
线性分类器:假设数据特征与分类目标之间是线性关系的模型,通过累加计算每个维度的特征与各自权重的乘积来帮助类别决策。
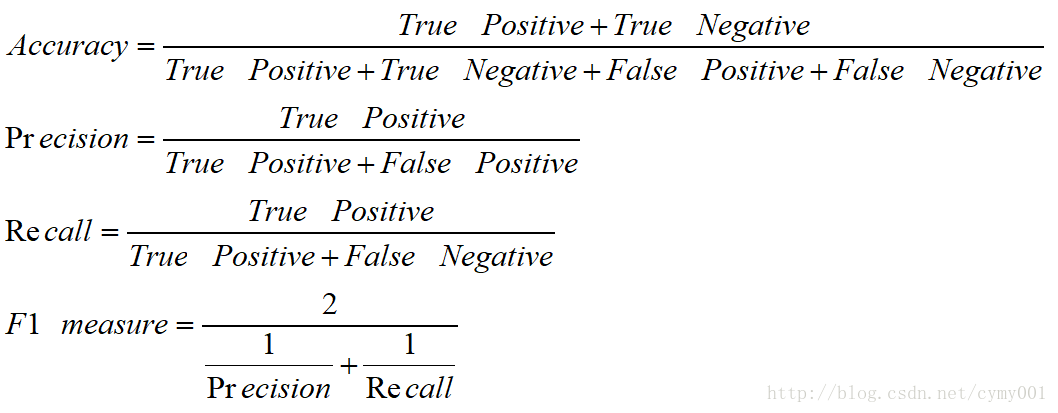
F1 measure 是Precision和Recall两个指标的调和平均数,对于Precision和Recall更加接近的模型 F1 measure 的得分会更高。
import numpy as np
import pandas as pd
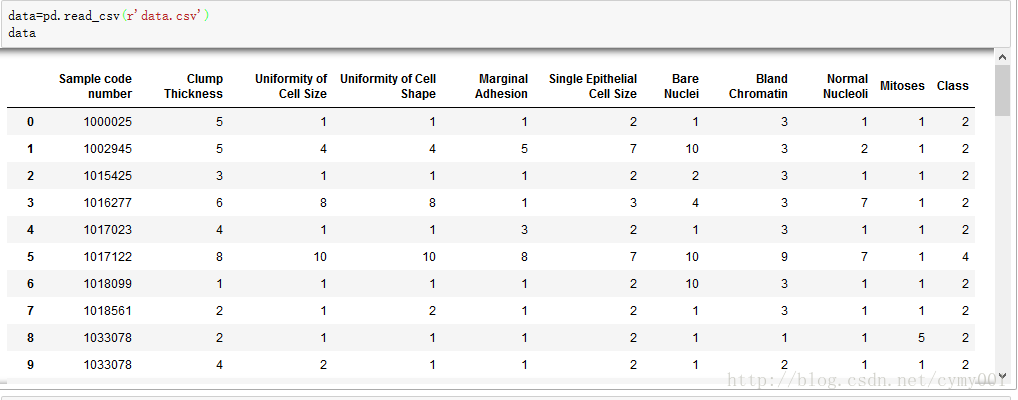
column_names=['Sample code number','Clump Thickness','Uniformity of Cell Size',
'Uniformity of Cell Shape','Marginal Adhesion',
'Single Epithelial Cell Size','Bare Nuclei','Bland Chromatin',
'Normal Nucleoli','Mitoses','Class']
data=pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/breast-cancer-wisconsin/breast-cancer-wisconsin.data',
names=column_names)
data.to_csv(r'data.csv',index=None)
data=pd.read_csv(r'data.csv')
datadata=data.replace(to_replace='?',value=np.nan) #将?替换为标准缺失值
data=data.dropna(how='any') #丢弃带有缺失值的数据样本
data.shape
#Output:(683, 11)from distutils.version import LooseVersion as Version
from sklearn import __version__ as sklearn_version
from sklearn import datasets
if Version(sklearn_version) < '0.18':
from sklearn.cross_validation import train_test_split
else:
from sklearn.model_selection import train_test_split
X_train,X_test,y_train,y_test = train_test_split(data[column_names[1:10]],data[column_names[10]], test_size=0.25, random_state=33)y_train.value_counts()
#Output:2 344
# 4 168
# Name: Class, dtype: int64y_test.value_counts()
#Output:2 100
# 4 71
# Name: Class, dtype: int64from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import LogisticRegression,SGDClassifier
ss=StandardScaler() #数据幅度标准化
X_train=ss.fit_transform(X_train)
X_test=ss.transform(X_test) #注意,对测试集不需要fit,用和训练集一样的变换lr=LogisticRegression() #初始化
lr.fit(X_train,y_train) #fit训练模型参数
lr_y_predict=lr.predict(X_test) #用训练好的模型lr进行预测,结果存储在变量lr_y_predict里
sgdc=SGDClassifier()
sgdc.fit(X_train,y_train)
sgdc_y_predict=sgdc.predict(X_test)from sklearn.metrics import classification_report #Accuracy,Precision,Recall,f1-score
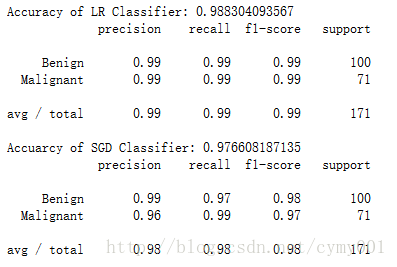
print('Accuracy of LR Classifier:',lr.score(X_test,y_test))
print(classification_report(y_test,lr_y_predict,target_names=['Benign','Malignant']))
print('Accuarcy of SGD Classifier:',sgdc.score(X_test,y_test))
print(classification_report(y_test,sgdc_y_predict,target_names=['Benign','Malignant']))