- Moodle + Websoft9:创新教育的强大组合,助力教学与学习
开源软件
Moodle+Websoft9:构建未来课堂的技术基石一、Moodle:开源生态的深度解析•模块化设计:支持超800个官方插件,如H5P交互内容创作、BigBlueButton虚拟课堂,满足个性化教学需求。•学习分析引擎:内置LearningAnalyticsAPI,可集成Python/R语言进行深度学习,预测学生学业风险。•移动优先战略:MoodleApp支持离线学习、扫码签到,2023年新增A
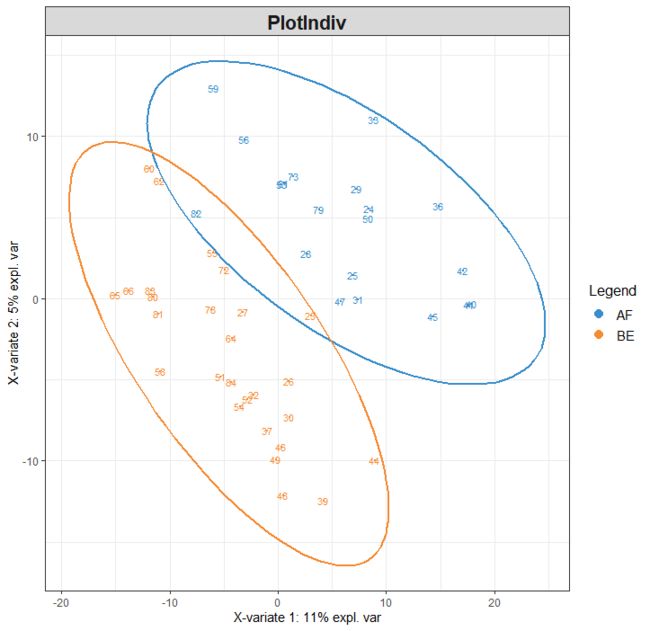
- R语言绘图 | 环状柱状图+散点柱状组合图绘制
小杜的生信筆記
R语言精美图形绘制教程r语言开发语言科研绘图生物信息学
原文:R语言绘图|环状柱状图+散点柱状组合图绘制(点击访问)小杜的生信筆記,主要发表或收录生物信息学教程,以及基于R分析和可视化(包括数据分析,图形绘制等);分享感兴趣的文献和学习资料!!
- AIGC带来数据革命:R语言如何成为数据科学家的秘密武器?
程序边界
AIGCr语言开发语言
文章目录一、R语言的基础特性1.1R语言的起源与发展1.2R语言的核心优势二、R语言在AIGC中的应用场景2.1数据预处理与清洗2.2文本分析与生成2.3机器学习与模型构建2.4数据可视化与报告生成三、R语言在AIGC中的具体案例3.1金融数据分析与预测3.2医疗数据分析与建模3.3社交媒体数据分析与情感分析四、R语言在AIGC中的未来展望4.1与深度学习框架的集成4.2与云计算平台的集成4.3与
- edger多组差异性分析_R语言统计分析微生物组数据
weixin_39961636
edger多组差异性分析
我在学习这本书记了一些笔记,如果你有学习,欢迎分享你的笔记或者教程。我的已有笔记汇总如下:宏基因组学习笔记宏基因组学习笔记2宏基因组笔记(第二章)R语言宏基因组学统计分析学习笔记(第三章-1)R语言宏基因组学统计分析学习笔记(第三章-2)https://link.springer.com/book/10.1007/978-981-13-1534-3下载方法,sci-hub大法啦。出版日期:2018
- 23章12节:抽样的蒙特卡洛方法
DAT|R科学与人工智能
用R探索医药数据科学r-4.2.1开发语言数据库人工智能r
蒙特卡洛方法作为一种基于随机抽样的数值计算技术,在工程、金融、统计、物理等众多领域中得到了广泛应用。该方法通过对大量随机数的模拟,来解决那些难以解析求解的问题。在实际问题中,常常需要从一个复杂分布中抽取样本,而传统的直接抽样方法可能难以实现。为了解决这一问题,接受‐拒绝抽样方法应运而生。本文旨在介绍如何利用R语言实现蒙特卡洛方法,特别是如何通过接受‐拒绝抽样从已知分布中抽取样本。文章以参数为(3,
- R语言使用table1包绘制(生成)三线表实战:单变量分列构建三线表、编写自定义函数在三线表中添加p值
statistics.insight
R语言入门课r语言数据挖掘机器学习
R语言使用table1包绘制(生成)三线表实战:单变量分列构建三线表、编写自定义函数在三线表中添加p值目录R语言使用table1包绘制(生成)三线表、使用单变量分列构建三线表、编写自定义函数在三线表中添加p值#三线表是什么?#导入包并构建仿真数据#R语言使用table1包绘制(生成)三线表、使用单变量分列构建三线表、编写自定义函数在三线表中添加p值#三线表是什么?三线表本来是微软公司的word编辑
- R语言的移动应用开发
滕若岚
包罗万象golang开发语言后端
R语言的移动应用开发在数据科学和统计分析的大潮中,R语言因其强大的数据处理和可视化能力而备受青睐。然而,R语言对移动应用开发的适用性并未得到广泛关注。本文将探讨R语言在移动应用开发中的潜力及其工具,并提供一些实践示例,希望能为读者在这一新兴领域的探索提供帮助。一、R语言概述R语言是一种用于统计计算和图形绘制的编程语言,因其简洁的语法和丰富的包生态系统受到数据科学家的广泛使用。R语言的强大之处在于其
- R语言:初始环境配置
一头大学牲
程序--编程记录r语言开发语言
文章目录R语言的配置URL和种子R语言的配置在R中安装languageserver包:(直接在R.exe中运行即可)install.packages("languageserver")关于jupyternotebook如何编写R语言:(好像每种jupyternotebook支持的编程语言,解释器都有对应的jupyternotebook)install.packages('IRkernel')IRk
- 【科研绘图系列】R语言绘制网络相关图(cor network plot)
生信学习者1
SCI科研绘图系列r语言数据分析数据挖掘数据可视化
禁止商业或二改转载,仅供自学使用,侵权必究,如需截取部分内容请后台联系作者!文章目录介绍加载R包数据下载导入数据数据预处理画图1画图2组合图形输出图片系统信息介绍【科研绘图系列】R语言绘制网络相关图(cornetworkplot)加载R包library(tidyverse)library(ggraph)library(igraph)library(patchwork)conflicted::
- ggplot2设置坐标轴范围_作图技巧018篇第二坐标轴在ggplot2中的实现
weixin_39618597
ggplot2设置坐标轴范围R语言ggplot2移除图例按键精灵定位坐标循环
“ggplot2中的次级坐标轴”生活科学哥-R语言科学2020-06-128:35在平时作图中,我们有时希望在一个坐标中进行二个坐标轴的设定,也是为了方便数据的显示。这个过程在EXCEL等当中比较容易实现,但是,如何在R中实现呢?今天我们就来讲一讲操作的过程。数据准备先准备如下数据:library(ggplot2)library(scales)library(magrittr)dfdata.fra
- 23章9节:分层随机抽样及其在R语言中的实现与验证
DAT|R科学与人工智能
用R探索医药数据科学r语言开发语言r-4.2.1机器学习人工智能算法
在统计学和数据科学的实际工作中,抽样方法始终扮演着至关重要的角色。如何从庞大的总体中获取具有代表性的样本,一直是数据分析过程中需要面对的核心问题之一。分层随机抽样作为一种常用的抽样方法,因其能够针对总体中的不同亚群体(层)进行有针对性的抽样,从而提高样本代表性、降低抽样误差,被广泛应用于社会调查、市场研究、医学试验等各个领域。本文旨在系统地阐述分层随机抽样的理论基础、抽样方法及其在R语言中的实现,
- R语言高效数据处理-自定义EXCEL数据排版
LEEBELOVED
r语言R语言高效处理数据excel批处理
注:以下代码均为实际数据处理中的笔记摘录,所以很零散1、自定义excel表数据输出格式、布局在实际数据处理中为了提升效率,将Excel报表交付给需求方时减少手动调整的环节很有必要#1.1设置表头格式header_style=0', style=sheet_style)#数据写入writeData(data_deal_main01,'data_deal_test1',data_de
- Java:AI 浪潮中的隐形支柱 —— 探秘 Java 在人工智能领域的独特地位
琢磨先生David
人工智能
引言在人工智能技术席卷全球的今天,当人们谈论AI开发时,Python、R语言、C++等工具总是最先被提及。然而在这个充满创新的领域,有一个"老兵"正悄然发挥着不可替代的作用——自1995年诞生至今的Java语言,凭借其独特的工程化基因,正在构建起AI世界的底层基础设施。本文将揭示Java如何在大数据、机器学习、企业级AI系统等领域持续创造价值。一、Java的AI基因解码跨平台优势的现代意义"一次编
- R 语言科研绘图第 31 期 --- 韦恩图-基础
TigerZ 生信宝库
r语言贴图程序人生开发语言
在发表科研论文的过程中,科研绘图是必不可少的,一张好看的图形会是文章很大的加分项。为了便于使用,本系列文章介绍的所有绘图都已收录到了sciRplot项目中,获取方式:R语言科研绘图模板---sciRplothttps://mp.weixin.qq.com/s/QA_8LVqjkdg4A16zLonw4w?payreadticket=HElUE5WWmBflodEFw10g0l2NrRotj8kbU
- 23章11节:自助抽样及其在R语言中的实现与验证
DAT|R科学与人工智能
用R探索医药数据科学r语言开发语言r-4.2.1microsoft信息可视化
在统计学中,数据分析的核心任务之一是如何在样本数据的基础上推断总体的性质。传统方法往往依赖于已知的概率分布假设和解析推导,但在现实问题中,我们往往无法准确得知总体分布,或者数据样本量较小,难以满足经典统计推断方法的要求。自助抽样作为一种非参数的计算方法,为我们提供了基于样本数据“自我重复”构建抽样分布的途径。1977年,斯坦福大学的B.Efron在著名论文《BootstrapMethods:Ano
- R语言将向量数据按照行方式转化为矩阵数据(设置参数byrow为TRUE)
sdgfbhgfj
R语言初见机器学习数据挖掘人工智能数据分析r语言
R语言将向量数据按照行方式转化为矩阵数据(设置参数byrow为TRUE)目录R语言将向量数据按照行方式转化为矩阵数据(设置参数byrow为TRUE)R语言是解决什么问题的?R语言将向量数据按照行方式转化为矩阵数据(设置参数byrow为TRUE)安利一个R语言的优秀博主及其CSDN专栏:R语言是解决什么问题的?R是一个有着统计分析功能及强大作图功能的软件系统,是由奥克兰大学统计学系的RossIhak
- R语言学习实例:使用R进行数据可视化
PixelCoder
信息可视化r语言学习R语言
R语言学习实例:使用R进行数据可视化R语言是一种功能强大且广泛使用的统计分析和数据可视化工具。在本实例中,我们将使用R语言来创建一些常见的数据可视化图表,包括散点图、柱状图和折线图。我们将展示如何使用R的基本绘图功能和一些常用的绘图库来生成这些图表。散点图是一种用于显示两个变量之间关系的图表。我们可以使用R的基本绘图功能来创建散点图。下面是一个示例代码,展示如何使用R创建散点图:#创建示例数据x<
- 探索R语言:经典案例解析与源代码
翠绿探寻
r语言信息可视化开发语言R语言
探索R语言:经典案例解析与源代码引言:R语言是一种流行的数据分析和统计建模工具,具有丰富的功能和广泛的应用领域。在本文中,我们将通过经典案例来探索R语言的一些重要功能和技术。我们将提供相应的源代码,以便读者能够实际运行并理解这些示例。案例一:数据导入与处理在数据分析中,数据导入和处理是首要任务。R语言提供了丰富的函数和包,用于处理各种数据格式。下面是一个简单的示例,演示了如何导入和处理CSV格式的
- R语言dataframe数据索引、访问: 使用attach函数绑定dataframe数据、这样可以直接使用列名称访问dataframe的列数据
omhdxgb
R语言123r语言数据挖掘人工智能机器学习数据分析
R语言dataframe数据索引、访问:使用attach函数绑定dataframe数据、这样可以直接使用列名称访问dataframe的列数据目录R语言dataframe数据索引、访问:使用attach函数绑定dataframe数据、这样可以直接使用列名称访问dataframe的列数据R语言特点R语言dataframe数据索引、访问:使用attach函数绑定dataframe数据、这样可以直接使用列
- R语言向量vector数据类型元素索引、访问:使用中括号[]和:符号以及乘法符号获取向量中指定范围内的偶数索引元素
omhdxgb
R语言123r语言机器学习数据挖掘人工智能数据分析
R语言向量vector数据类型元素索引、访问:使用中括号[]和:符号以及乘法符号获取向量中指定范围内的偶数索引元素目录R语言向量vector数据类型元素索引、访问:使用中括号[]和:符号以及乘法符号获取向量中指定范围内的偶数索引元素R语言特点R语言向量vector数据类型元素索引、访问:使用中括号[]和:符号以及乘法符号获取向量中指定范围内的偶数索引元素R可以在CRAN(Comprehensive
- R语言安装github包出现的错误,object "XXX" is not exported by "namespace:viridisLite"
momo酱豆是沃
anaconda各种问题
自己遇上了类似的问题,当时是把所有导致这个问题出现时安装的所有包我都卸载了,再次重装的。弄了很久,我发现都是在安装各种包让我更新我不更新导致的后果R,告诉我一个道理,一定要听话,让你更新就更新,不然我的bug出到让你崩溃。下图借用以为博主的图(https://blog.csdn.net/yw_vine/article/details/79631042)原连接R语言安装github包出现的错误,ob
- R语言 ggplot2 可视化生成高分辨率图片实战
PixelEnigma
r语言开发语言R语言
R语言ggplot2可视化生成高分辨率图片实战在数据分析和可视化领域,R语言一直是研究人员和数据科学家们的首选工具。其中,ggplot2包是R语言中最受欢迎和强大的可视化工具之一。它提供了许多灵活且精美的图形选项,使用户能够轻松创建具有吸引力和信息丰富的图表。本文将介绍如何使用ggplot2包在R语言中生成高分辨率的图片。我们将探索不同的保存选项,以确保我们获得清晰、适应各种输出需求的图像。首先,
- Java 中操作 R:深度整合与高效应用
froginwe11
开发语言
Java中操作R:深度整合与高效应用引言随着大数据和机器学习的快速发展,R语言在数据分析和可视化方面扮演着越来越重要的角色。而Java作为一种广泛应用于企业级应用开发的语言,其强大的功能和稳定性使其成为构建高性能应用的首选。本文将探讨Java如何操作R语言,实现高效的数据分析应用。一、Java操作R的背景R语言优势:R语言拥有丰富的统计分析、数据可视化工具和机器学习算法库,是数据分析领域的首选语言
- R语言对高频交易订单流进行建模分析 4
oxuzhenyi
实验楼课程机器学习R
一、实验介绍--订单流模型拟合1.1实验知识点指数核hawkes过程拟合正反馈强度分析订单量影响分析1.2实验环境R3.4.1Rstudio二、订单流模型拟合在上节中我们对订单流数据做了一些统计分析,对交易的一些特征有了一些粗浅的理解,在本节中我们要做的是利用实际数据来拟合hawkes过程,看一看真实数据的订单流动力学中有什么特征。首先我们仍是选出交易时间内的数据:library(tidyvers
- R语言对高频交易订单流进行建模分析 3
oxuzhenyi
实验楼课程机器学习R
一、实验介绍--订单流数据描述分析1.1实验知识点订单流数据表示订单间隔分析订单信息率平稳性研究订单流动性研究限价单相对价格分析1.2实验环境R3.4.1Rstudio二、订单流数据描述分析2.1订单流数据表示当我们在金融市场上做交易时,可以看到一个委托单簿,上面陈列着买价和卖价以及它们对应的量,举个例子,比特币市场的订单簿:可以看到红色代表的是卖价,或者说是ask,而绿色代表的是买价,或者说是b
- 【自学笔记】R语言基础知识点总览-持续更新
Long_poem
笔记r语言开发语言
提示:文章写完后,目录可以自动生成,如何生成可参考右边的帮助文档文章目录R语言基础知识点总览1.R语言简介2.R语言安装与环境配置3.R语言基础语法3.1数据类型3.2向量与矩阵3.3数据框与列表4.控制结构4.1条件语句4.2循环结构5.函数6.数据可视化总结R语言基础知识点总览1.R语言简介R是一种用于统计计算和图形的编程语言和软件环境。R语言由RossIhaka和RobertGentlema
- R语言 决策树、svm支持向量机、随机森林
别叫我名字20
R语言决策树支持向量机r语言
本人正在学习R语言,想利用这个平台记录自己一些自己的学习情况,方便以后查找,也想分享出来提供一些资料给同样学习R语言的同学们。(如果内容有错误,欢迎大家批评指正)1.决策树我们使用的还是RStudio自带的数据集iris。#######################决策树模型install.packages("rpart")#安装库library("rpart")dt<-function(dat
- 决策树、朴素贝叶斯、随机森林、支持向量机、XGBoost 和 LightGBM算法的R语言实现
生信与基因组学
生信分析项目进阶技能合集算法机器学习r语言
基本逻辑(1)使用rnorm函数生成5个特征变量x1到x5,并根据这些特征变量的线性组合生成一个二分类的响应变量y;(2)将生成的数据存储在数据框中,处理缺失值,并将响应变量转换为因子类型;(3)使用决策树、朴素贝叶斯、随机森林、支持向量机、XGBoost和LightGBM六种机器学习模型算法对数据进行训练和评估;(4)将各个模型的准确率和AUC值存储在结果数据框中,并通过柱状图展示结果。1.R包
- R语言2——数据类型和基本运算
朝荣
#R语言R语言运算R数据类型
R语言2——数据类型和基本运算目录R语言2——数据类型和基本运算1.R的数据类型1.R的数据类型(1)Logical(逻辑型):只有两个值TRUE,FALSE(2)Numeric(数字):整数、小数等(3)Complex(复合型):带有虚数i的数,如1+2i(4)Character(字符):包含在“”之中,如“hello!”(5)Vectors向量:c()函数,将元素组合成一个向量。c(1,2,3
- R语言获取数据——手工输入数据
蜗牛数据分析
R语言从入门到实战r语言开发语言
在R语言中获取数据集的方法有多种,例如读取Excel文件、数据库中的文件,而当我们没有这些渠道能够获取到数据集时,也可以手工输入数据,即通过键盘输入数据,它是获取数据集的最简单方法。另外,还可以在代码中直接输入数据,下面分别进行介绍。数据编辑器R提供了内置的数据编辑器,通过edit()函数调用该编辑器就可以实现手工输入数据。举例1:通过数据编辑器创建学生成绩表下面实现“学生成绩表”,具体步骤如下:
- java线程的无限循环和退出
3213213333332132
java
最近想写一个游戏,然后碰到有关线程的问题,网上查了好多资料都没满足。
突然想起了前段时间看的有关线程的视频,于是信手拈来写了一个线程的代码片段。
希望帮助刚学java线程的童鞋
package thread;
import java.text.SimpleDateFormat;
import java.util.Calendar;
import java.util.Date
- tomcat 容器
BlueSkator
tomcatWebservlet
Tomcat的组成部分 1、server
A Server element represents the entire Catalina servlet container. (Singleton) 2、service
service包括多个connector以及一个engine,其职责为处理由connector获得的客户请求。
3、connector
一个connector
- php递归,静态变量,匿名函数使用
dcj3sjt126com
PHP递归函数匿名函数静态变量引用传参
<!doctype html>
<html lang="en">
<head>
<meta charset="utf-8">
<title>Current To-Do List</title>
</head>
<body>
- 属性颜色字体变化
周华华
JavaScript
function changSize(className){
var diva=byId("fot")
diva.className=className;
}
</script>
<style type="text/css">
.max{
background: #900;
color:#039;
- 将properties内容放置到map中
g21121
properties
代码比较简单:
private static Map<Object, Object> map;
private static Properties p;
static {
//读取properties文件
InputStream is = XXX.class.getClassLoader().getResourceAsStream("xxx.properti
- [简单]拼接字符串
53873039oycg
字符串
工作中遇到需要从Map里面取值拼接字符串的情况,自己写了个,不是很好,欢迎提出更优雅的写法,代码如下:
import java.util.HashMap;
import java.uti
- Struts2学习
云端月影
最近开始关注struts2的新特性,从这个版本开始,Struts开始使用convention-plugin代替codebehind-plugin来实现struts的零配置。
配置文件精简了,的确是简便了开发过程,但是,我们熟悉的配置突然disappear了,真是一下很不适应。跟着潮流走吧,看看该怎样来搞定convention-plugin。
使用Convention插件,你需要将其JAR文件放
- Java新手入门的30个基本概念二
aijuans
java新手java 入门
基本概念: 1.OOP中唯一关系的是对象的接口是什么,就像计算机的销售商她不管电源内部结构是怎样的,他只关系能否给你提供电就行了,也就是只要知道can or not而不是how and why.所有的程序是由一定的属性和行为对象组成的,不同的对象的访问通过函数调用来完成,对象间所有的交流都是通过方法调用,通过对封装对象数据,很大限度上提高复用率。 2.OOP中最重要的思想是类,类是模板是蓝图,
- jedis 简单使用
antlove
javarediscachecommandjedis
jedis.RedisOperationCollection.java
package jedis;
import org.apache.log4j.Logger;
import redis.clients.jedis.Jedis;
import java.util.List;
import java.util.Map;
import java.util.Set;
pub
- PL/SQL的函数和包体的基础
百合不是茶
PL/SQL编程函数包体显示包的具体数据包
由于明天举要上课,所以刚刚将代码敲了一遍PL/SQL的函数和包体的实现(单例模式过几天好好的总结下再发出来);以便明天能更好的学习PL/SQL的循环,今天太累了,所以早点睡觉,明天继续PL/SQL总有一天我会将你永远的记载在心里,,,
函数;
函数:PL/SQL中的函数相当于java中的方法;函数有返回值
定义函数的
--输入姓名找到该姓名的年薪
create or re
- Mockito(二)--实例篇
bijian1013
持续集成mockito单元测试
学习了基本知识后,就可以实战了,Mockito的实际使用还是比较麻烦的。因为在实际使用中,最常遇到的就是需要模拟第三方类库的行为。
比如现在有一个类FTPFileTransfer,实现了向FTP传输文件的功能。这个类中使用了a
- 精通Oracle10编程SQL(7)编写控制结构
bijian1013
oracle数据库plsql
/*
*编写控制结构
*/
--条件分支语句
--简单条件判断
DECLARE
v_sal NUMBER(6,2);
BEGIN
select sal into v_sal from emp
where lower(ename)=lower('&name');
if v_sal<2000 then
update emp set
- 【Log4j二】Log4j属性文件配置详解
bit1129
log4j
如下是一个log4j.properties的配置
log4j.rootCategory=INFO, stdout , R
log4j.appender.stdout=org.apache.log4j.ConsoleAppender
log4j.appender.stdout.layout=org.apache.log4j.PatternLayout
log4j.appe
- java集合排序笔记
白糖_
java
public class CollectionDemo implements Serializable,Comparable<CollectionDemo>{
private static final long serialVersionUID = -2958090810811192128L;
private int id;
private String nam
- java导致linux负载过高的定位方法
ronin47
定位java进程ID
可以使用top或ps -ef |grep java
![图片描述][1]
根据进程ID找到最消耗资源的java pid
比如第一步找到的进程ID为5431
执行
top -p 5431 -H
![图片描述][2]
打印java栈信息
$ jstack -l 5431 > 5431.log
在栈信息中定位具体问题
将消耗资源的Java PID转
- 给定能随机生成整数1到5的函数,写出能随机生成整数1到7的函数
bylijinnan
函数
import java.util.ArrayList;
import java.util.List;
import java.util.Random;
public class RandNFromRand5 {
/**
题目:给定能随机生成整数1到5的函数,写出能随机生成整数1到7的函数。
解法1:
f(k) = (x0-1)*5^0+(x1-
- PL/SQL Developer保存布局
Kai_Ge
近日由于项目需要,数据库从DB2迁移到ORCAL,因此数据库连接客户端选择了PL/SQL Developer。由于软件运用不熟悉,造成了很多麻烦,最主要的就是进入后,左边列表有很多选项,自己删除了一些选项卡,布局很满意了,下次进入后又恢复了以前的布局,很是苦恼。在众多PL/SQL Developer使用技巧中找到如下这段:
&n
- [未来战士计划]超能查派[剧透,慎入]
comsci
计划
非常好看,超能查派,这部电影......为我们这些热爱人工智能的工程技术人员提供一些参考意见和思想........
虽然电影里面的人物形象不是非常的可爱....但是非常的贴近现实生活....
&nbs
- Google Map API V2
dai_lm
google map
以后如果要开发包含google map的程序就更麻烦咯
http://www.cnblogs.com/mengdd/archive/2013/01/01/2841390.html
找到篇不错的文章,大家可以参考一下
http://blog.sina.com.cn/s/blog_c2839d410101jahv.html
1. 创建Android工程
由于v2的key需要G
- java数据计算层的几种解决方法2
datamachine
javasql集算器
2、SQL
SQL/SP/JDBC在这里属于一类,这是老牌的数据计算层,性能和灵活性是它的优势。但随着新情况的不断出现,单纯用SQL已经难以满足需求,比如: JAVA开发规模的扩大,数据量的剧增,复杂计算问题的涌现。虽然SQL得高分的指标不多,但都是权重最高的。
成熟度:5星。最成熟的。
- Linux下Telnet的安装与运行
dcj3sjt126com
linuxtelnet
Linux下Telnet的安装与运行 linux默认是使用SSH服务的 而不安装telnet服务 如果要使用telnet 就必须先安装相应的软件包 即使安装了软件包 默认的设置telnet 服务也是不运行的 需要手工进行设置 如果是redhat9,则在第三张光盘中找到 telnet-server-0.17-25.i386.rpm
- PHP中钩子函数的实现与认识
dcj3sjt126com
PHP
假如有这么一段程序:
function fun(){
fun1();
fun2();
}
首先程序执行完fun1()之后执行fun2()然后fun()结束。
但是,假如我们想对函数做一些变化。比如说,fun是一个解析函数,我们希望后期可以提供丰富的解析函数,而究竟用哪个函数解析,我们希望在配置文件中配置。这个时候就可以发挥钩子的力量了。
我们可以在fu
- EOS中的WorkSpace密码修改
蕃薯耀
修改WorkSpace密码
EOS中BPS的WorkSpace密码修改
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
蕃薯耀 201
- SpringMVC4零配置--SpringSecurity相关配置【SpringSecurityConfig】
hanqunfeng
SpringSecurity
SpringSecurity的配置相对来说有些复杂,如果是完整的bean配置,则需要配置大量的bean,所以xml配置时使用了命名空间来简化配置,同样,spring为我们提供了一个抽象类WebSecurityConfigurerAdapter和一个注解@EnableWebMvcSecurity,达到同样减少bean配置的目的,如下:
applicationContex
- ie 9 kendo ui中ajax跨域的问题
jackyrong
AJAX跨域
这两天遇到个问题,kendo ui的datagrid,根据json去读取数据,然后前端通过kendo ui的datagrid去渲染,但很奇怪的是,在ie 10,ie 11,chrome,firefox等浏览器中,同样的程序,
浏览起来是没问题的,但把应用放到公网上的一台服务器,
却发现如下情况:
1) ie 9下,不能出现任何数据,但用IE 9浏览器浏览本机的应用,却没任何问题
- 不要让别人笑你不能成为程序员
lampcy
编程程序员
在经历六个月的编程集训之后,我刚刚完成了我的第一次一对一的编码评估。但是事情并没有如我所想的那般顺利。
说实话,我感觉我的脑细胞像被轰炸过一样。
手慢慢地离开键盘,心里很压抑。不禁默默祈祷:一切都会进展顺利的,对吧?至少有些地方我的回答应该是没有遗漏的,是不是?
难道我选择编程真的是一个巨大的错误吗——我真的永远也成不了程序员吗?
我需要一点点安慰。在自我怀疑,不安全感和脆弱等等像龙卷风一
- 马皇后的贤德
nannan408
马皇后不怕朱元璋的坏脾气,并敢理直气壮地吹耳边风。众所周知,朱元璋不喜欢女人干政,他认为“后妃虽母仪天下,然不可使干政事”,因为“宠之太过,则骄恣犯分,上下失序”,因此还特地命人纂述《女诫》,以示警诫。但马皇后是个例外。
有一次,马皇后问朱元璋道:“如今天下老百姓安居乐业了吗?”朱元璋不高兴地回答:“这不是你应该问的。”马皇后振振有词地回敬道:“陛下是天下之父,
- 选择某个属性值最大的那条记录(不仅仅包含指定属性,而是想要什么属性都可以)
Rainbow702
sqlgroup by最大值max最大的那条记录
好久好久不写SQL了,技能退化严重啊!!!
直入主题:
比如我有一张表,file_info,
它有两个属性(但实际不只,我这里只是作说明用):
file_code, file_version
同一个code可能对应多个version
现在,我想针对每一个code,取得它相关的记录中,version 值 最大的那条记录,
SQL如下:
select
*
- VBScript脚本语言
tntxia
VBScript
VBScript 是基于VB的脚本语言。主要用于Asp和Excel的编程。
VB家族语言简介
Visual Basic 6.0
源于BASIC语言。
由微软公司开发的包含协助开发环境的事
- java中枚举类型的使用
xiao1zhao2
javaenum枚举1.5新特性
枚举类型是j2se在1.5引入的新的类型,通过关键字enum来定义,常用来存储一些常量.
1.定义一个简单的枚举类型
public enum Sex {
MAN,
WOMAN
}
枚举类型本质是类,编译此段代码会生成.class文件.通过Sex.MAN来访问Sex中的成员,其返回值是Sex类型.
2.常用方法
静态的values()方