作者:桂。
时间:2017-03-06 20:57:22
链接:http://www.cnblogs.com/xingshansi/p/6511916.html
前言
本文为Hilbert变换一篇的内容补充,主要内容为:
1)EMD原理介绍
2)代码分析
3)一种权衡的小trick
4)问题补充
内容主要为自己的学习总结,并多有借鉴他人,最后一并给出链接。
一、EMD原理介绍
A-EMD的意义
很多人都知道EMD(Empirical Mode Decomposition)可以将信号分解不同频率特性,并且结合Hilbert求解包络以及瞬时频率。EMD、Hilbert、瞬时频率三者有无内在联系?答案是:有。
按照Hilbert变换一篇的介绍,
$f(t) = \frac{{d\Phi (t)}}{{d(t)}}$
然而,这样求解瞬时频率在某些情况下有问题,可能出现$f(t)$为负的情况:我1秒手指动5下,频率是5Hz;反过来,频率为8Hz时,手指1秒动8下,可如果频率为-5Hz呢?负频率没有意义。
考虑信号
$x(t) = {x_1}(t) + {x_2}(t) = {A_1}{e^{j{\omega _1}t}} + {A_2}{e^{j{\omega _2}t}} = A(t){e^{j\varphi (t)}}$
为了简单起见,假设$A_1$和$A_2$恒定,且$\omega_1$和$\omega_2$是正的。信号$x(t)$的频谱应由两个在$\omega_1$和$\omega_2$的$\delta$函数组成,即
$X(\omega ) = {A_1}\delta (\omega - {\omega _1}) + {A_2}\delta (\omega - {\omega _2})$
因为假设$\omega_1$和$\omega_2$是正的,所以该信号解析。求得相位
$\Phi (t) = \frac{{{A_1}\sin {\omega _1}t + {A_{\rm{2}}}\sin {\omega _{\rm{2}}}t}}{{{A_1}\cos {\omega _1}t + {A_{\rm{2}}}\cos {\omega _{\rm{2}}}t}}$
分别取两组参数,对$t$求导,得到对应参数下的瞬时频率:
参数:
$\omega_1 = 10Hz$和$\omega_2 = 20Hz$.
- 组1:{$A_1 = 0.2, A_2 = 1$};
- 组2:{$A_1 = 1.2, A_2 = 1$}
对于组2,瞬时频率出现了负值。
可见:
对任意信号进行Hilbert变换,可能出现无法解释、缺乏实际意义的频率分量。Norden E. Hung等人对瞬时频率进行研究后发现,只有满足特定条件的信号,其瞬时频率才具有物理意义,并将此类信号成为:IMF/基本模式分量。
B-EMD基本原理
此处给一个原理图:
C-基本模式分量(IMF)
EMD分解的IMF其瞬时频率具有实际物理意义,原因有两点:
- 限定1:
- 在整个数据序列中,极值点的数量$N_e$(包括极大值、极小值点)与过零点的数量必须相等,或最多相差1个,即$(N_e-1) \le N_e \ge (N_e+1)$.
- 限定2:
- 在任意时间点$t_i$上,信号局部极大值确定的上包络线$f_{max}(t)$和局部极小值确定的下包络线$f_{min}(t)$的均值为0.
限定1即要求信号具有类似传统平稳高斯过程的分布;限定2要求局部均值为0,同时用局部最大、最小值的包络作为近似,从而信号局部对称,避免了不对称带来的瞬时频率波动。
D-VMD
关于VMD(Variational Mode Decomposition),具体原理可以参考其论文,这里我们只要记住一点:其分解的各个基本分量——即各解析信号的瞬时频率具有实际的物理意义。
二、代码分析
首先给出信号分别用VMD、EMD的分解结果:
给出对应的代码:
%--------------- Preparation
clear all;
close all;
clc;
% Time Domain 0 to T
T = 1000;
fs = 1/T;
t = (1:T)/T;
freqs = 2*pi*(t-0.5-1/T)/(fs);
% center frequencies of components
f_1 = 2;
f_2 = 24;
f_3 = 288;
% modes
v_1 = (cos(2*pi*f_1*t));
v_2 = 1/4*(cos(2*pi*f_2*t));
v_3 = 1/16*(cos(2*pi*f_3*t));
% for visualization purposes
wsub{1} = 2*pi*f_1;
wsub{2} = 2*pi*f_2;
wsub{3} = 2*pi*f_3;
% composite signal, including noise
f = v_1 + v_2 + v_3 + 0.1*randn(size(v_1));
% some sample parameters for VMD
alpha = 2000; % moderate bandwidth constraint
tau = 0; % noise-tolerance (no strict fidelity enforcement)
K = 4; % 4 modes
DC = 0; % no DC part imposed
init = 1; % initialize omegas uniformly
tol = 1e-7;
%--------------- Run actual VMD code
[u, u_hat, omega] = VMD(f, alpha, tau, K, DC, init, tol);
subplot(size(u,1)+1,2,1);
plot(t,f,'k');grid on;
title('VMD分解');
subplot(size(u,1)+1,2,2);
plot(freqs,abs(fft(f)),'k');grid on;
title('对应频谱');
for i = 2:size(u,1)+1
subplot(size(u,1)+1,2,i*2-1);
plot(t,u(i-1,:),'k');grid on;
subplot(size(u,1)+1,2,i*2);
plot(freqs,abs(fft(u(i-1,:))),'k');grid on;
end
%---------------run EMD code
imf = emd(f);
figure;
subplot(size(imf,1)+1,2,1);
plot(t,f,'k');grid on;
title('EMD分解');
subplot(size(imf,1)+1,2,2);
plot(freqs,abs(fft(f)),'k');grid on;
title('对应频谱');
for i = 2:size(imf,1)+1
subplot(size(imf,1)+1,2,i*2-1);
plot(t,imf(i-1,:),'k');grid on;
subplot(size(imf,1)+1,2,i*2);
plot(freqs,abs(fft(imf(i-1,:))),'k');grid on;
end
附上两个子程序的code.
VMD:
function [u, u_hat, omega] = VMD(signal, alpha, tau, K, DC, init, tol)
% Variational Mode Decomposition
% Authors: Konstantin Dragomiretskiy and Dominique Zosso
% zosso@math.ucla.edu --- http://www.math.ucla.edu/~zosso
% Initial release 2013-12-12 (c) 2013
%
% Input and Parameters:
% ---------------------
% signal - the time domain signal (1D) to be decomposed
% alpha - the balancing parameter of the data-fidelity constraint
% tau - time-step of the dual ascent ( pick 0 for noise-slack )
% K - the number of modes to be recovered
% DC - true if the first mode is put and kept at DC (0-freq)
% init - 0 = all omegas start at 0
% 1 = all omegas start uniformly distributed
% 2 = all omegas initialized randomly
% tol - tolerance of convergence criterion; typically around 1e-6
%
% Output:
% -------
% u - the collection of decomposed modes
% u_hat - spectra of the modes
% omega - estimated mode center-frequencies
%
% When using this code, please do cite our paper:
% -----------------------------------------------
% K. Dragomiretskiy, D. Zosso, Variational Mode Decomposition, IEEE Trans.
% on Signal Processing (in press)
% please check here for update reference:
% http://dx.doi.org/10.1109/TSP.2013.2288675
%---------- Preparations
% Period and sampling frequency of input signal
save_T = length(signal);
fs = 1/save_T;
% extend the signal by mirroring
T = save_T;
f_mirror(1:T/2) = signal(T/2:-1:1);
f_mirror(T/2+1:3*T/2) = signal;
f_mirror(3*T/2+1:2*T) = signal(T:-1:T/2+1);
f = f_mirror;
% Time Domain 0 to T (of mirrored signal)
T = length(f);
t = (1:T)/T;
% Spectral Domain discretization
freqs = t-0.5-1/T;
% Maximum number of iterations (if not converged yet, then it won't anyway)
N = 500;
% For future generalizations: individual alpha for each mode
Alpha = alpha*ones(1,K);
% Construct and center f_hat
f_hat = fftshift((fft(f)));
f_hat_plus = f_hat;
f_hat_plus(1:T/2) = 0;
% matrix keeping track of every iterant // could be discarded for mem
u_hat_plus = zeros(N, length(freqs), K);
% Initialization of omega_k
omega_plus = zeros(N, K);
switch init
case 1
for i = 1:K
omega_plus(1,i) = (0.5/K)*(i-1);
end
case 2
omega_plus(1,:) = sort(exp(log(fs) + (log(0.5)-log(fs))*rand(1,K)));
otherwise
omega_plus(1,:) = 0;
end
% if DC mode imposed, set its omega to 0
if DC
omega_plus(1,1) = 0;
end
% start with empty dual variables
lambda_hat = zeros(N, length(freqs));
% other inits
uDiff = tol+eps; % update step
n = 1; % loop counter
sum_uk = 0; % accumulator
% ----------- Main loop for iterative updates
while ( uDiff > tol && n < N ) % not converged and below iterations limit
% update first mode accumulator
k = 1;
sum_uk = u_hat_plus(n,:,K) + sum_uk - u_hat_plus(n,:,1);
% update spectrum of first mode through Wiener filter of residuals
u_hat_plus(n+1,:,k) = (f_hat_plus - sum_uk - lambda_hat(n,:)/2)./(1+Alpha(1,k)*(freqs - omega_plus(n,k)).^2);
% update first omega if not held at 0
if ~DC
omega_plus(n+1,k) = (freqs(T/2+1:T)*(abs(u_hat_plus(n+1, T/2+1:T, k)).^2)')/sum(abs(u_hat_plus(n+1,T/2+1:T,k)).^2);
end
% update of any other mode
for k=2:K
% accumulator
sum_uk = u_hat_plus(n+1,:,k-1) + sum_uk - u_hat_plus(n,:,k);
% mode spectrum
u_hat_plus(n+1,:,k) = (f_hat_plus - sum_uk - lambda_hat(n,:)/2)./(1+Alpha(1,k)*(freqs - omega_plus(n,k)).^2);
% center frequencies
omega_plus(n+1,k) = (freqs(T/2+1:T)*(abs(u_hat_plus(n+1, T/2+1:T, k)).^2)')/sum(abs(u_hat_plus(n+1,T/2+1:T,k)).^2);
end
% Dual ascent
lambda_hat(n+1,:) = lambda_hat(n,:) + tau*(sum(u_hat_plus(n+1,:,:),3) - f_hat_plus);
% loop counter
n = n+1;
% converged yet?
uDiff = eps;
for i=1:K
uDiff = uDiff + 1/T*(u_hat_plus(n,:,i)-u_hat_plus(n-1,:,i))*conj((u_hat_plus(n,:,i)-u_hat_plus(n-1,:,i)))';
end
uDiff = abs(uDiff);
end
%------ Postprocessing and cleanup
% discard empty space if converged early
N = min(N,n);
omega = omega_plus(1:N,:);
% Signal reconstruction
u_hat = zeros(T, K);
u_hat((T/2+1):T,:) = squeeze(u_hat_plus(N,(T/2+1):T,:));
u_hat((T/2+1):-1:2,:) = squeeze(conj(u_hat_plus(N,(T/2+1):T,:)));
u_hat(1,:) = conj(u_hat(end,:));
u = zeros(K,length(t));
for k = 1:K
u(k,:)=real(ifft(ifftshift(u_hat(:,k))));
end
% remove mirror part
u = u(:,T/4+1:3*T/4);
% recompute spectrum
clear u_hat;
for k = 1:K
u_hat(:,k)=fftshift(fft(u(k,:)))';
end
end
EMD:
%EMD computes Empirical Mode Decomposition
%
%
% Syntax
%
%
% IMF = EMD(X)
% IMF = EMD(X,...,'Option_name',Option_value,...)
% IMF = EMD(X,OPTS)
% [IMF,ORT,NB_ITERATIONS] = EMD(...)
%
%
% Description
%
%
% IMF = EMD(X) where X is a real vector computes the Empirical Mode
% Decomposition [1] of X, resulting in a matrix IMF containing 1 IMF per row, the
% last one being the residue. The default stopping criterion is the one proposed
% in [2]:
%
% at each point, mean_amplitude < THRESHOLD2*envelope_amplitude
% &
% mean of boolean array {(mean_amplitude)/(envelope_amplitude) > THRESHOLD} < TOLERANCE
% &
% |#zeros-#extrema|<=1
%
% where mean_amplitude = abs(envelope_max+envelope_min)/2
% and envelope_amplitude = abs(envelope_max-envelope_min)/2
%
% IMF = EMD(X) where X is a complex vector computes Bivariate Empirical Mode
% Decomposition [3] of X, resulting in a matrix IMF containing 1 IMF per row, the
% last one being the residue. The default stopping criterion is similar to the
% one proposed in [2]:
%
% at each point, mean_amplitude < THRESHOLD2*envelope_amplitude
% &
% mean of boolean array {(mean_amplitude)/(envelope_amplitude) > THRESHOLD} < TOLERANCE
%
% where mean_amplitude and envelope_amplitude have definitions similar to the
% real case
%
% IMF = EMD(X,...,'Option_name',Option_value,...) sets options Option_name to
% the specified Option_value (see Options)
%
% IMF = EMD(X,OPTS) is equivalent to the above syntax provided OPTS is a struct
% object with field names corresponding to option names and field values being the
% associated values
%
% [IMF,ORT,NB_ITERATIONS] = EMD(...) returns an index of orthogonality
% ________
% _ |IMF(i,:).*IMF(j,:)|
% ORT = \ _____________________
% /
% ? || X ||?% i~=j
%
% and the number of iterations to extract each mode in NB_ITERATIONS
%
%
% Options
%
%
% stopping criterion options:
%
% STOP: vector of stopping parameters [THRESHOLD,THRESHOLD2,TOLERANCE]
% if the input vector's length is less than 3, only the first parameters are
% set, the remaining ones taking default values.
% default: [0.05,0.5,0.05]
%
% FIX (int): disable the default stopping criterion and do exactly
% number of sifting iterations for each mode
%
% FIX_H (int): disable the default stopping criterion and do sifting
% iterations with |#zeros-#extrema|<=1 to stop [4]
%
% bivariate/complex EMD options:
%
% COMPLEX_VERSION: selects the algorithm used for complex EMD ([3])
% COMPLEX_VERSION = 1: "algorithm 1"
% COMPLEX_VERSION = 2: "algorithm 2" (default)
%
% NDIRS: number of directions in which envelopes are computed (default 4)
% rem: the actual number of directions (according to [3]) is 2*NDIRS
%
% other options:
%
% T: sampling times (line vector) (default: 1:length(x))
%
% MAXITERATIONS: maximum number of sifting iterations for the computation of each
% mode (default: 2000)
%
% MAXMODES: maximum number of imfs extracted (default: Inf)
%
% DISPLAY: if equals to 1 shows sifting steps with pause
% if equals to 2 shows sifting steps without pause (movie style)
% rem: display is disabled when the input is complex
%
% INTERP: interpolation scheme: 'linear', 'cubic', 'pchip' or 'spline' (default)
% see interp1 documentation for details
%
% MASK: masking signal used to improve the decomposition according to [5]
%
%
% Examples
%
%
%X = rand(1,512);
%
%IMF = emd(X);
%
%IMF = emd(X,'STOP',[0.1,0.5,0.05],'MAXITERATIONS',100);
%
%T=linspace(0,20,1e3);
%X = 2*exp(i*T)+exp(3*i*T)+.5*T;
%IMF = emd(X,'T',T);
%
%OPTIONS.DISLPAY = 1;
%OPTIONS.FIX = 10;
%OPTIONS.MAXMODES = 3;
%[IMF,ORT,NBITS] = emd(X,OPTIONS);
%
%
% References
%
%
% [1] N. E. Huang et al., "The empirical mode decomposition and the
% Hilbert spectrum for non-linear and non stationary time series analysis",
% Proc. Royal Soc. London A, Vol. 454, pp. 903-995, 1998
%
% [2] G. Rilling, P. Flandrin and P. Gon鏰lves
% "On Empirical Mode Decomposition and its algorithms",
% IEEE-EURASIP Workshop on Nonlinear Signal and Image Processing
% NSIP-03, Grado (I), June 2003
%
% [3] G. Rilling, P. Flandrin, P. Gon鏰lves and J. M. Lilly.,
% "Bivariate Empirical Mode Decomposition",
% Signal Processing Letters (submitted)
%
% [4] N. E. Huang et al., "A confidence limit for the Empirical Mode
% Decomposition and Hilbert spectral analysis",
% Proc. Royal Soc. London A, Vol. 459, pp. 2317-2345, 2003
%
% [5] R. Deering and J. F. Kaiser, "The use of a masking signal to improve
% empirical mode decomposition", ICASSP 2005
%
%
% See also
% emd_visu (visualization),
% emdc, emdc_fix (fast implementations of EMD),
% cemdc, cemdc_fix, cemdc2, cemdc2_fix (fast implementations of bivariate EMD),
% hhspectrum (Hilbert-Huang spectrum)
%
%
% G. Rilling, last modification: 3.2007
% gabriel.rilling@ens-lyon.fr
function [imf,ort,nbits] = emd(varargin)
[x,t,sd,sd2,tol,MODE_COMPLEX,ndirs,display_sifting,sdt,sd2t,r,imf,k,nbit,NbIt,MAXITERATIONS,FIXE,FIXE_H,MAXMODES,INTERP,mask] = init(varargin{:});
if display_sifting
fig_h = figure;
end
%main loop : requires at least 3 extrema to proceed
while ~stop_EMD(r,MODE_COMPLEX,ndirs) && (k < MAXMODES+1 || MAXMODES == 0) && ~any(mask)
% current mode
m = r;
% mode at previous iteration
mp = m;
%computation of mean and stopping criterion
if FIXE
[stop_sift,moyenne] = stop_sifting_fixe(t,m,INTERP,MODE_COMPLEX,ndirs);
elseif FIXE_H
stop_count = 0;
[stop_sift,moyenne] = stop_sifting_fixe_h(t,m,INTERP,stop_count,FIXE_H,MODE_COMPLEX,ndirs);
else
[stop_sift,moyenne] = stop_sifting(m,t,sd,sd2,tol,INTERP,MODE_COMPLEX,ndirs);
end
% in case the current mode is so small that machine precision can cause
% spurious extrema to appear
if (max(abs(m))) < (1e-10)*(max(abs(x)))
if ~stop_sift
warning('emd:warning','forced stop of EMD : too small amplitude')
else
disp('forced stop of EMD : too small amplitude')
end
break
end
% sifting loop
while ~stop_sift && nbitMAXITERATIONS/5 && mod(nbit,floor(MAXITERATIONS/10))==0 && ~FIXE && nbit > 100)
disp(['mode ',int2str(k),', iteration ',int2str(nbit)])
if exist('s','var')
disp(['stop parameter mean value : ',num2str(s)])
end
[im,iM] = extr(m);
disp([int2str(sum(m(im) > 0)),' minima > 0; ',int2str(sum(m(iM) < 0)),' maxima < 0.'])
end
%sifting
m = m - moyenne;
%computation of mean and stopping criterion
if FIXE
[stop_sift,moyenne] = stop_sifting_fixe(t,m,INTERP,MODE_COMPLEX,ndirs);
elseif FIXE_H
[stop_sift,moyenne,stop_count] = stop_sifting_fixe_h(t,m,INTERP,stop_count,FIXE_H,MODE_COMPLEX,ndirs);
else
[stop_sift,moyenne,s] = stop_sifting(m,t,sd,sd2,tol,INTERP,MODE_COMPLEX,ndirs);
end
% display
if display_sifting && ~MODE_COMPLEX
NBSYM = 2;
[indmin,indmax] = extr(mp);
[tmin,tmax,mmin,mmax] = boundary_conditions(indmin,indmax,t,mp,mp,NBSYM);
envminp = interp1(tmin,mmin,t,INTERP);
envmaxp = interp1(tmax,mmax,t,INTERP);
envmoyp = (envminp+envmaxp)/2;
if FIXE || FIXE_H
display_emd_fixe(t,m,mp,r,envminp,envmaxp,envmoyp,nbit,k,display_sifting)
else
sxp=2*(abs(envmoyp))./(abs(envmaxp-envminp));
sp = mean(sxp);
display_emd(t,m,mp,r,envminp,envmaxp,envmoyp,s,sp,sxp,sdt,sd2t,nbit,k,display_sifting,stop_sift)
end
end
mp = m;
nbit=nbit+1;
NbIt=NbIt+1;
if(nbit==(MAXITERATIONS-1) && ~FIXE && nbit > 100)
if exist('s','var')
warning('emd:warning',['forced stop of sifting : too many iterations... mode ',int2str(k),'. stop parameter mean value : ',num2str(s)])
else
warning('emd:warning',['forced stop of sifting : too many iterations... mode ',int2str(k),'.'])
end
end
end % sifting loop
imf(k,:) = m;
if display_sifting
disp(['mode ',int2str(k),' stored'])
end
nbits(k) = nbit;
k = k+1;
r = r - m;
nbit=0;
end %main loop
if any(r) && ~any(mask)
imf(k,:) = r;
end
ort = io(x,imf);
if display_sifting
close
end
end
%---------------------------------------------------------------------------------------------------
% tests if there are enough (3) extrema to continue the decomposition
function stop = stop_EMD(r,MODE_COMPLEX,ndirs)
if MODE_COMPLEX
for k = 1:ndirs
phi = (k-1)*pi/ndirs;
[indmin,indmax] = extr(real(exp(i*phi)*r));
ner(k) = length(indmin) + length(indmax);
end
stop = any(ner < 3);
else
[indmin,indmax] = extr(r);
ner = length(indmin) + length(indmax);
stop = ner < 3;
end
end
%---------------------------------------------------------------------------------------------------
% computes the mean of the envelopes and the mode amplitude estimate
function [envmoy,nem,nzm,amp] = mean_and_amplitude(m,t,INTERP,MODE_COMPLEX,ndirs)
NBSYM = 2;
if MODE_COMPLEX
switch MODE_COMPLEX
case 1
for k = 1:ndirs
phi = (k-1)*pi/ndirs;
y = real(exp(-i*phi)*m);
[indmin,indmax,indzer] = extr(y);
nem(k) = length(indmin)+length(indmax);
nzm(k) = length(indzer);
[tmin,tmax,zmin,zmax] = boundary_conditions(indmin,indmax,t,y,m,NBSYM);
envmin(k,:) = interp1(tmin,zmin,t,INTERP);
envmax(k,:) = interp1(tmax,zmax,t,INTERP);
end
envmoy = mean((envmin+envmax)/2,1);
if nargout > 3
amp = mean(abs(envmax-envmin),1)/2;
end
case 2
for k = 1:ndirs
phi = (k-1)*pi/ndirs;
y = real(exp(-i*phi)*m);
[indmin,indmax,indzer] = extr(y);
nem(k) = length(indmin)+length(indmax);
nzm(k) = length(indzer);
[tmin,tmax,zmin,zmax] = boundary_conditions(indmin,indmax,t,y,y,NBSYM);
envmin(k,:) = exp(i*phi)*interp1(tmin,zmin,t,INTERP);
envmax(k,:) = exp(i*phi)*interp1(tmax,zmax,t,INTERP);
end
envmoy = mean((envmin+envmax),1);
if nargout > 3
amp = mean(abs(envmax-envmin),1)/2;
end
end
else
[indmin,indmax,indzer] = extr(m);
nem = length(indmin)+length(indmax);
nzm = length(indzer);
[tmin,tmax,mmin,mmax] = boundary_conditions(indmin,indmax,t,m,m,NBSYM);
envmin = interp1(tmin,mmin,t,INTERP);
envmax = interp1(tmax,mmax,t,INTERP);
envmoy = (envmin+envmax)/2;
if nargout > 3
amp = mean(abs(envmax-envmin),1)/2;
end
end
end
%-------------------------------------------------------------------------------
% default stopping criterion
function [stop,envmoy,s] = stop_sifting(m,t,sd,sd2,tol,INTERP,MODE_COMPLEX,ndirs)
try
[envmoy,nem,nzm,amp] = mean_and_amplitude(m,t,INTERP,MODE_COMPLEX,ndirs);
sx = abs(envmoy)./amp;
s = mean(sx);
stop = ~((mean(sx > sd) > tol | any(sx > sd2)) & (all(nem > 2)));
if ~MODE_COMPLEX
stop = stop && ~(abs(nzm-nem)>1);
end
catch
stop = 1;
envmoy = zeros(1,length(m));
s = NaN;
end
end
%-------------------------------------------------------------------------------
% stopping criterion corresponding to option FIX
function [stop,moyenne]= stop_sifting_fixe(t,m,INTERP,MODE_COMPLEX,ndirs)
try
moyenne = mean_and_amplitude(m,t,INTERP,MODE_COMPLEX,ndirs);
stop = 0;
catch
moyenne = zeros(1,length(m));
stop = 1;
end
end
%-------------------------------------------------------------------------------
% stopping criterion corresponding to option FIX_H
function [stop,moyenne,stop_count]= stop_sifting_fixe_h(t,m,INTERP,stop_count,FIXE_H,MODE_COMPLEX,ndirs)
try
[moyenne,nem,nzm] = mean_and_amplitude(m,t,INTERP,MODE_COMPLEX,ndirs);
if (all(abs(nzm-nem)>1))
stop = 0;
stop_count = 0;
else
stop_count = stop_count+1;
stop = (stop_count == FIXE_H);
end
catch
moyenne = zeros(1,length(m));
stop = 1;
end
end
%-------------------------------------------------------------------------------
% displays the progression of the decomposition with the default stopping criterion
function display_emd(t,m,mp,r,envmin,envmax,envmoy,s,sb,sx,sdt,sd2t,nbit,k,display_sifting,stop_sift)
subplot(4,1,1)
plot(t,mp);hold on;
plot(t,envmax,'--k');plot(t,envmin,'--k');plot(t,envmoy,'r');
title(['IMF ',int2str(k),'; iteration ',int2str(nbit),' before sifting']);
set(gca,'XTick',[])
hold off
subplot(4,1,2)
plot(t,sx)
hold on
plot(t,sdt,'--r')
plot(t,sd2t,':k')
title('stop parameter')
set(gca,'XTick',[])
hold off
subplot(4,1,3)
plot(t,m)
title(['IMF ',int2str(k),'; iteration ',int2str(nbit),' after sifting']);
set(gca,'XTick',[])
subplot(4,1,4);
plot(t,r-m)
title('residue');
disp(['stop parameter mean value : ',num2str(sb),' before sifting and ',num2str(s),' after'])
if stop_sift
disp('last iteration for this mode')
end
if display_sifting == 2
pause(0.01)
else
pause
end
end
%---------------------------------------------------------------------------------------------------
% displays the progression of the decomposition with the FIX and FIX_H stopping criteria
function display_emd_fixe(t,m,mp,r,envmin,envmax,envmoy,nbit,k,display_sifting)
subplot(3,1,1)
plot(t,mp);hold on;
plot(t,envmax,'--k');plot(t,envmin,'--k');plot(t,envmoy,'r');
title(['IMF ',int2str(k),'; iteration ',int2str(nbit),' before sifting']);
set(gca,'XTick',[])
hold off
subplot(3,1,2)
plot(t,m)
title(['IMF ',int2str(k),'; iteration ',int2str(nbit),' after sifting']);
set(gca,'XTick',[])
subplot(3,1,3);
plot(t,r-m)
title('residue');
if display_sifting == 2
pause(0.01)
else
pause
end
end
%---------------------------------------------------------------------------------------
% defines new extrema points to extend the interpolations at the edges of the
% signal (mainly mirror symmetry)
function [tmin,tmax,zmin,zmax] = boundary_conditions(indmin,indmax,t,x,z,nbsym)
lx = length(x);
if (length(indmin) + length(indmax) < 3)
error('not enough extrema')
end
% boundary conditions for interpolations :
if indmax(1) < indmin(1)
if x(1) > x(indmin(1))
lmax = fliplr(indmax(2:min(end,nbsym+1)));
lmin = fliplr(indmin(1:min(end,nbsym)));
lsym = indmax(1);
else
lmax = fliplr(indmax(1:min(end,nbsym)));
lmin = [fliplr(indmin(1:min(end,nbsym-1))),1];
lsym = 1;
end
else
if x(1) < x(indmax(1))
lmax = fliplr(indmax(1:min(end,nbsym)));
lmin = fliplr(indmin(2:min(end,nbsym+1)));
lsym = indmin(1);
else
lmax = [fliplr(indmax(1:min(end,nbsym-1))),1];
lmin = fliplr(indmin(1:min(end,nbsym)));
lsym = 1;
end
end
if indmax(end) < indmin(end)
if x(end) < x(indmax(end))
rmax = fliplr(indmax(max(end-nbsym+1,1):end));
rmin = fliplr(indmin(max(end-nbsym,1):end-1));
rsym = indmin(end);
else
rmax = [lx,fliplr(indmax(max(end-nbsym+2,1):end))];
rmin = fliplr(indmin(max(end-nbsym+1,1):end));
rsym = lx;
end
else
if x(end) > x(indmin(end))
rmax = fliplr(indmax(max(end-nbsym,1):end-1));
rmin = fliplr(indmin(max(end-nbsym+1,1):end));
rsym = indmax(end);
else
rmax = fliplr(indmax(max(end-nbsym+1,1):end));
rmin = [lx,fliplr(indmin(max(end-nbsym+2,1):end))];
rsym = lx;
end
end
tlmin = 2*t(lsym)-t(lmin);
tlmax = 2*t(lsym)-t(lmax);
trmin = 2*t(rsym)-t(rmin);
trmax = 2*t(rsym)-t(rmax);
% in case symmetrized parts do not extend enough
if tlmin(1) > t(1) || tlmax(1) > t(1)
if lsym == indmax(1)
lmax = fliplr(indmax(1:min(end,nbsym)));
else
lmin = fliplr(indmin(1:min(end,nbsym)));
end
if lsym == 1
error('bug')
end
lsym = 1;
tlmin = 2*t(lsym)-t(lmin);
tlmax = 2*t(lsym)-t(lmax);
end
if trmin(end) < t(lx) || trmax(end) < t(lx)
if rsym == indmax(end)
rmax = fliplr(indmax(max(end-nbsym+1,1):end));
else
rmin = fliplr(indmin(max(end-nbsym+1,1):end));
end
if rsym == lx
error('bug')
end
rsym = lx;
trmin = 2*t(rsym)-t(rmin);
trmax = 2*t(rsym)-t(rmax);
end
zlmax =z(lmax);
zlmin =z(lmin);
zrmax =z(rmax);
zrmin =z(rmin);
tmin = [tlmin t(indmin) trmin];
tmax = [tlmax t(indmax) trmax];
zmin = [zlmin z(indmin) zrmin];
zmax = [zlmax z(indmax) zrmax];
end
%---------------------------------------------------------------------------------------------------
%extracts the indices of extrema
function [indmin, indmax, indzer] = extr(x,t)
if(nargin==1)
t=1:length(x);
end
m = length(x);
if nargout > 2
x1=x(1:m-1);
x2=x(2:m);
indzer = find(x1.*x2<0);
if any(x == 0)
iz = find( x==0 );
indz = [];
if any(diff(iz)==1)
zer = x == 0;
dz = diff([0 zer 0]);
debz = find(dz == 1);
finz = find(dz == -1)-1;
indz = round((debz+finz)/2);
else
indz = iz;
end
indzer = sort([indzer indz]);
end
end
d = diff(x);
n = length(d);
d1 = d(1:n-1);
d2 = d(2:n);
indmin = find(d1.*d2<0 & d1<0)+1;
indmax = find(d1.*d2<0 & d1>0)+1;
% when two or more successive points have the same value we consider only one extremum in the middle of the constant area
% (only works if the signal is uniformly sampled)
if any(d==0)
imax = [];
imin = [];
bad = (d==0);
dd = diff([0 bad 0]);
debs = find(dd == 1);
fins = find(dd == -1);
if debs(1) == 1
if length(debs) > 1
debs = debs(2:end);
fins = fins(2:end);
else
debs = [];
fins = [];
end
end
if length(debs) > 0
if fins(end) == m
if length(debs) > 1
debs = debs(1:(end-1));
fins = fins(1:(end-1));
else
debs = [];
fins = [];
end
end
end
lc = length(debs);
if lc > 0
for k = 1:lc
if d(debs(k)-1) > 0
if d(fins(k)) < 0
imax = [imax round((fins(k)+debs(k))/2)];
end
else
if d(fins(k)) > 0
imin = [imin round((fins(k)+debs(k))/2)];
end
end
end
end
if length(imax) > 0
indmax = sort([indmax imax]);
end
if length(imin) > 0
indmin = sort([indmin imin]);
end
end
end
%---------------------------------------------------------------------------------------------------
function ort = io(x,imf)
% ort = IO(x,imf) computes the index of orthogonality
%
% inputs : - x : analyzed signal
% - imf : empirical mode decomposition
n = size(imf,1);
s = 0;
for i = 1:n
for j =1:n
if i~=j
s = s + abs(sum(imf(i,:).*conj(imf(j,:)))/sum(x.^2));
end
end
end
ort = 0.5*s;
end
%---------------------------------------------------------------------------------------------------
function [x,t,sd,sd2,tol,MODE_COMPLEX,ndirs,display_sifting,sdt,sd2t,r,imf,k,nbit,NbIt,MAXITERATIONS,FIXE,FIXE_H,MAXMODES,INTERP,mask] = init(varargin)
x = varargin{1};
if nargin == 2
if isstruct(varargin{2})
inopts = varargin{2};
else
error('when using 2 arguments the first one is the analyzed signal X and the second one is a struct object describing the options')
end
elseif nargin > 2
try
inopts = struct(varargin{2:end});
catch
error('bad argument syntax')
end
end
% default for stopping
defstop = [0.05,0.5,0.05];
opt_fields = {'t','stop','display','maxiterations','fix','maxmodes','interp','fix_h','mask','ndirs','complex_version'};
defopts.stop = defstop;
defopts.display = 0;
defopts.t = 1:max(size(x));
defopts.maxiterations = 2000;
defopts.fix = 0;
defopts.maxmodes = 0;
defopts.interp = 'spline';
defopts.fix_h = 0;
defopts.mask = 0;
defopts.ndirs = 4;
defopts.complex_version = 2;
opts = defopts;
if(nargin==1)
inopts = defopts;
elseif nargin == 0
error('not enough arguments')
end
names = fieldnames(inopts);
for nom = names'
if ~any(strcmpi(char(nom), opt_fields))
error(['bad option field name: ',char(nom)])
end
if ~isempty(eval(['inopts.',char(nom)])) % empty values are discarded
eval(['opts.',lower(char(nom)),' = inopts.',char(nom),';'])
end
end
t = opts.t;
stop = opts.stop;
display_sifting = opts.display;
MAXITERATIONS = opts.maxiterations;
FIXE = opts.fix;
MAXMODES = opts.maxmodes;
INTERP = opts.interp;
FIXE_H = opts.fix_h;
mask = opts.mask;
ndirs = opts.ndirs;
complex_version = opts.complex_version;
if ~isvector(x)
error('X must have only one row or one column')
end
if size(x,1) > 1
x = x.';
end
if ~isvector(t)
error('option field T must have only one row or one column')
end
if ~isreal(t)
error('time instants T must be a real vector')
end
if size(t,1) > 1
t = t';
end
if (length(t)~=length(x))
error('X and option field T must have the same length')
end
if ~isvector(stop) || length(stop) > 3
error('option field STOP must have only one row or one column of max three elements')
end
if ~all(isfinite(x))
error('data elements must be finite')
end
if size(stop,1) > 1
stop = stop';
end
L = length(stop);
if L < 3
stop(3)=defstop(3);
end
if L < 2
stop(2)=defstop(2);
end
if ~ischar(INTERP) || ~any(strcmpi(INTERP,{'linear','cubic','spline'}))
error('INTERP field must be ''linear'', ''cubic'', ''pchip'' or ''spline''')
end
%special procedure when a masking signal is specified
if any(mask)
if ~isvector(mask) || length(mask) ~= length(x)
error('masking signal must have the same dimension as the analyzed signal X')
end
if size(mask,1) > 1
mask = mask.';
end
opts.mask = 0;
imf1 = emd(x+mask,opts);
imf2 = emd(x-mask,opts);
if size(imf1,1) ~= size(imf2,1)
warning('emd:warning',['the two sets of IMFs have different sizes: ',int2str(size(imf1,1)),' and ',int2str(size(imf2,1)),' IMFs.'])
end
S1 = size(imf1,1);
S2 = size(imf2,1);
if S1 ~= S2
if S1 < S2
tmp = imf1;
imf1 = imf2;
imf2 = tmp;
end
imf2(max(S1,S2),1) = 0;
end
imf = (imf1+imf2)/2;
end
sd = stop(1);
sd2 = stop(2);
tol = stop(3);
lx = length(x);
sdt = sd*ones(1,lx);
sd2t = sd2*ones(1,lx);
if FIXE
MAXITERATIONS = FIXE;
if FIXE_H
error('cannot use both ''FIX'' and ''FIX_H'' modes')
end
end
MODE_COMPLEX = ~isreal(x)*complex_version;
if MODE_COMPLEX && complex_version ~= 1 && complex_version ~= 2
error('COMPLEX_VERSION parameter must equal 1 or 2')
end
% number of extrema and zero-crossings in residual
ner = lx;
nzr = lx;
r = x;
if ~any(mask) % if a masking signal is specified "imf" already exists at this stage
imf = [];
end
k = 1;
% iterations counter for extraction of 1 mode
nbit=0;
% total iterations counter
NbIt=0;
end
%---------------------------------------------------------------------------------------------------
关于EMD,有对应的工具箱。VMD也有扩展的二维分解,此处不再展开。
三、一种权衡的小trick
关于瞬时频率的原理以及代码,参考另一篇博文。
比较来看:
- EMD分解的IMF分量个数不能人为设定,而VMD(Variational Mode Decomposition)则可以;
- 但VMD也有弊端:分解过多,则信号断断续续,没有多少规律可言。
能不能取长补短呢?
自己之前做了一个小code,放在这里,供大家交流使用(此理论为自己首创,版权所有,拿去也不介意!(●'◡'●))。
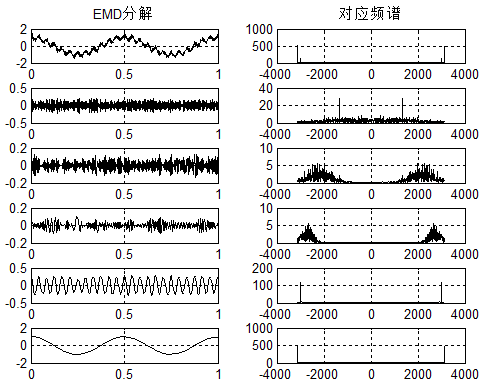
给定一个信号,下图是EMD分解结果,分解出了5个分量。
再来一个VMD(设定分量个数为3)的分解结果:
比较两个结果,可以发现:VMD的低频分量,更容易表达出经济波动的大趋势,而EMD则不易观察该特性。
或许有人会说:几个EMD分量叠加一下,也会有该效果,但如果不观察分解的数据,如何确定几个分量相加呢?更何况EMD总的IMF个数也是未知!
VMD的优势观察到了,但如何确定分量个数呢?
再来一个效果图:
这里分析了VMD分量从1~9,9种情况下某特征的曲线,可以观察到:个数增加到一定数量,曲线有了明显的下弯曲现象(该特性容易借助曲率,进行量化分析,不再展开),这个临界的个数就是分解的合适数量,此处:K=3,因为到4就有了明显的下弯曲。
可见通过该特征,即可理论上得出最优K。下面讲一讲这个某特征为何物?
上一段代码:
for st=1:9
K=st+1;
[u, u_hat, omega] = VMD(data, length(data), 0, K, 0, 1, 1e-5);
u=flipud(u);
resf=zeros(1,K);
for i=1:K
testdata=u(i,:);
hilbert(testdata');
z=hilbert(testdata'); % 希尔伯特变换
a=abs(z); % 包络线
fnor=instfreq(z); % 瞬时频率
resf(i)=mean(fnor);
end
subplot(3,3,st)
plot(resf,'k');title(['个数为',num2str(st)]);grid on;
end
没错,该特征就是:分量瞬时频率的均值。如果分解个数过大,则分量会出现断断絮絮地现象,特别是在高频,这样一来,即使是高频,平均瞬时频率反而低一些,这也是下弯曲的根本原因。
这个小trick就介绍到这里。
四、问题补充
HHT算法中,有两处存在端点效应,VMD是否也有呢?这一点没有再去验证。另外,关于Hilbert的端点效应,在另一篇博文已经给出。
参考:
宋知用:《MATLAB在语音信号分析和合成中的应用》
了凡春秋: http://blog.sina.com.cn/s/blog_6163bdeb0102e2cd.html
VMD-code:https://cn.mathworks.com/matlabcentral/fileexchange/44765-variational-mode-decomposition
EMD原理图:http://blog.sciencenet.cn/blog-244606-256958.html