R语言与属性数据分析
文章目录
- 1. 属性数据的描述性分析
- 1)属性数据形式的转化
- 2)属性数据的描述性统计图
- 3)属性数据的基本统计量
- 2. 单一属性分类数据
- 1)分类数据的概率检验
- 3. 四格表
- 1)四格表的独立性检验
- 2)四格表的边缘齐性(对称性)检验
- 4. 二维列联表
- 1)二维列联表的独立性检验
- 2)二维列联表的相合性度量和检验
- 3)方表的一致性度量和检验
- 5. 三维列联表
- 1)三维列联表的独立性检验
- 2)三维列联表的相合性检验
- 3)三维列联表的相关性检验
- 6. Logistic回归模型
- 1)二分类Logistic回归案例
1. 属性数据的描述性分析
1)属性数据形式的转化
* 将数据框中的分类数据(统计好的)转换成频数表
# input data
Reason<-c("操作","设备","工具","工艺","材料")
Count<-c(22,526,292,89,47)
data0<-data.frame(Reason,Count);data0
# change to the table
ftable<-table(rep(Reason,Count));data1
names(Count)<-Reason;Count
* 将数据框中的分类数据(未统计好的)转换成频数表
ftable<-table(data$classvar);ftable
* 频数表转换成频率表
ftable/sum(ftable)
prop.table(ftable)
* 列联表和概率列联表
ctable<-table(data);ctable
prop.table(ctable)
* 将频率表转化为数据集
Indicator<-c(1:5) // c('A','B','C')
data<-rep(Indicator,ftable);data
2)属性数据的描述性统计图
barplot(Count,main='条形图')
pie(Count,radius = 1,main='饼图')
library(qcc)
pareto.chart(Count, ylab = "频数",ylab2 = "累计百分比(%)",main='帕累托图')
3)属性数据的基本统计量
* 描述中心位置的统计量
median()
quantile( , probs = c(0.25,0.75))
getmode()
# the mode function
getmode<-function(x){
tab<-table(x)
tab.max<-max(tab)
if (all(tab == tab.max)) {mod<-NA}
else if(is.numeric(x)) {mod<-as.numeric(names(tab)[tab == tab.max])}
else {mod<-names(tab)[tab == tab.max]}
names(mod)<-'mode'
return(mod)
}
* 描述离散程度的统计量(离异比率、GS指数、熵)
var.ratio<-function(x){
tab<-table(x)
mod<-getmode(x)
ratio=1-tab[names(tab)==mod]/sum(tab)
names(ratio)<-'variation ratio'
return(ratio)
}
GS.index<-function(x){
tab<-table(x)
p<-prop.table(tab)
index<-1-sum(p^2)
names(index)<-'GS index'
return(index)
}
entropy<-function(x){
#x is the raw data
tab<-table(x)
p<-prop.table(tab)
entropy<- (-1)*sum(p*log(p))
names(entropy)<-'entropy'
return(entropy)
}
2. 单一属性分类数据
1)分类数据的概率检验
* 不含参数的Chi2检验
chisq.test(table, correct = TURE/FALSE, p = p0, rescale.p = TRUE/FALSE)
* 不含参数的LRT检验
library(RVAideMemoire)
G.test(table,p=p0)
# 等概率检验时,p0 = rep(1/length(x2), length(x2))
---------------------------------------------------------------
* 参数的极大似然估计(MLE) (用数值方法解)
* 含一个参数
fn_likelihood<-function(p){
... #log-likelihood function
}
result<-optimize(f=fn_likelihood, interval=c(0,1), maximum = TRUE);result
# optimize函数得到maximum和objective两个值
p_hat<-result$maximum # the MLE for p
p<-c(...)
* 含多个参数
fn_likelihood<-function(x){
p<-x[1]
q<-x[2]
... #log-likelihood function
}
result<-optim(par = c(0.1,0.1),fn_likelihood,control = list(fnscale = -1));result
p_hat<-result$par[1] # the MLE for p
q_hat<-result$par[2] # the MLE for q
---------------------------------------------------------------
* 含参数的Chi2检验
Chistat<-chisq.test(x, correct = FALSE, p = p, rescale.p = FALSE)$statistic
* 含参数的LRT检验
LRTstat<-G.test(x,p = p)$statistic
---------------------------------------------------------------
* 统计决策
# critical value
qchisq(p=0.95, df=r-m-1,lower.tail = TRUE)
# p value
pchisq(Chistat,df=r-m-1,lower.tail = FALSE)
pchisq(LRTstat,df=r-m-1,lower.tail = FALSE)
3. 四格表
1)四格表的独立性检验
* U检验(单边)
U.stat=sqrt(n)*(n11*n22-n12*n21)/sqrt(n1_*n2_*n_1*n_2) #未修正
U.stat=sqrt(n)*(n11*n22-n12*n21 ± n/2)/sqrt(n1_*n2_*n_1*n_2) #修正
pvalue=pnorm(U.stat, mean = 0, sd = 1, lower.tail = FALSE)
* Chi2检验(双边)
Chi.stat=n*(n11*n22-n12*n21)^2/(n1_*n2_*n_1*n_2) #未修正
Chi.stat=n*(abs(n11*n22-n12*n21)-n/2)^2/(n1_*n2_*n_1*n_2) #修正
pvalue=pchisq(Chi.stat, df=1, ncp=0, lower.tail = FALSE)
* LRT检验
LRT.stat1=-2*(n11*log(n1_*n_1/n11/n)+n12*log(n1_*n_2/n12/n)+n21*log(n2_*n_1/n21/n)+n22*log(n2_*n_2/n22/n))
pvalue=pchisq(LRT.stat, df=1, ncp=0, lower.tail = FALSE)
* Fisher精确检验
table<-matrix(c(n11,n12,n21,n22),byrow=TRUE,nrow=2,ncol=2)
fisher.test(x=table,alternative = "greater"/"less"/"two.sided")
* U近似精确检验
U.stat=sqrt(n-1)*(n11*n22-n12*n21)/sqrt(n1_*n2_*n_1*n_2) #未修正
U.stat=sqrt(n)*(n11*n22-n12*n21 ± n/2)/sqrt(n1_*n2_*n_1*n_2) #修正
* Mantel Haenszel Chi2近似精确检验
Chi.stat=(n-1)*(n11*n22-n12*n21)^2/(n1_*n2_*n_1*n_2) #未修正
Chi.stat=(n-1)*(abs(n11*n22-n12*n21)-n/2)^2/(n1_*n2_*n_1*n_2) #修正
* OR优比检验
U.stat <-log(n11*n22/(n12*n21))/sqrt(1/n11+1/n12+1/n21+1/n22)
Chi.stat <- log(n11*n22/(n12*n21))^2/(1/n11+1/n12+1/n21+1/n22)
2)四格表的边缘齐性(对称性)检验
* McNemar Chi2检验
Chi.stat=(n12-n21)^2/(n12+n21)
* LRT检验
LRT.stat=-2*(n12*log((n12+n21)/2/n12)+n21*log((n12+n21)/2/n21))
chisq.test 和 G.test 函数不适用于四格表的分析,但可以用在分类数据和列联表的检验中;
可用于四格表检验的自带函数只有fisher.test
4. 二维列联表
1)二维列联表的独立性检验
table<-matrix(c(...),byrow=TRUE,nrow=r,ncol=c)
chisq.test(table, correct = TURE/FALSE)
library(RVAideMemoire)
G.test(table)
2)二维列联表的相合性度量和检验
一定是单边检验
# Calculate Kendall_tau, gamma and Somers d
# x is a table(matrix)
assocoef<-function(x){
Gcount<-function(x){#this function is used to calculate G
r<-dim(x)[1];c<-dim(x)[2];G<-0
for (i in 1:(r-1)){
for (j in 1:(c-1)){
G<-G+x[i,j]*sum(x[(i+1):r,(j+1):c])
}
}
return(G)
}
Hcount<-function(x){#this function is used to calculate H
r<-dim(x)[1];c<-dim(x)[2];H<-0
for (i in 1:(r-1)){
for (j in 2:c){
H<-H+x[i,j]*sum(x[(i+1):r,1:(j-1)])
}
}
return(H)
}
TAcount<-function(x){#this function is used to calculate TA
r<-dim(x)[1];TA<-0
for (i in 1:r){
TA<-TA+sum(x[i,])*(sum(x[i,])-1)/2
}
return(TA)
}
TBcount<-function(x){#this function is used to calculate TB
c<-dim(x)[2];TB<-0
for (j in 1:c){
TB<-TB+sum(x[,j])*(sum(x[,j])-1)/2
}
return(TB)
}
G<-Gcount(x);H<-Hcount(x);TA<-TAcount(x);TB<-TBcount(x);n<-sum(x)
tau<-(G-H)/sqrt((n*(n-1)/2-TA)*(n*(n-1)/2-TB))
gamma<-(G-H)/(G+H)
dCR<-(G-H)/(n*(n-1)/2-TA)
dRC<-(G-H)/(n*(n-1)/2-TB)
return(list(G=G,H=H,tau=tau,gamma=gamma,dCR=dCR,dRC=dRC))
}
assocoef(table)
tau
gamma
dCR
dRC
# Association test statistic based on z=G-H (use the approximate variance for z)
assotest<-function(x){
G<-assocoef(x)$G; H<-assocoef(x)$H;n<-sum(x);z<-G-H
z.var<-(n^3-sum((rowSums(x))^3))*(n^3-sum((colSums(x))^3))/9/n^3
U<-z/sqrt(z.var)
chisqstat<-U^2
return(list(U=U,chisqstat=chisqstat,z=z,z.var=z.var))
}
assotest(table)
3)方表的一致性度量和检验
单边检验
# kappa and agreement test statistic
kappatest<-function(x){
n<-sum(x)
q1<-sum(diag(x))/n
q2<-sum(rowSums(x)*colSums(x))/n^2
kappa<-(q1-q2)/(1-q2)
var.kappa<-(q2+q2^2-sum(rowSums(x)*colSums(x)*(rowSums(x)+colSums(x)))/n^3)/(n-1)/(1-q2)^2
U<-kappa/sqrt(var.kappa)
return(list(kappa=kappa,var.kappa=var.kappa,U=U))
}
kappatest(table)
5. 三维列联表
1)三维列联表的独立性检验
- indtset3d 函数定义
- type = mutual.ind:完全独立性检验
- type = joint.ind.A | B | C:分块独立性检验
- type = cond.ind.A | B | C:条件独立性检验
indtest3d<-function(x,type="mutual.ind"){
#this function is for independence test for 3-dimension r*c*t contingency table
#x is an array including frequencies of r*c*t contingency table for row A,column B,stratum C.
#output includes Pearson chi-square test and likelihood ratio test
r<-dim(x)[1];c<-dim(x)[2];t<-dim(x)[3];n<-sum(x)
m<-array(0,dim=c(r,c,t))
Pearson.chisq<-likelihood.ratio.base<-0
if (type=="mutual.ind"){#for mutual independence (A,B,C)
for (i in 1:r){
for (j in 1:c){
for (k in 1:t){
m[i,j,k]<-sum(x[i,,])*sum(x[,j,])*sum(x[,,k])/n^2
Pearson.chisq<-Pearson.chisq+(x[i,j,k]-m[i,j,k])^2/m[i,j,k]
likelihood.ratio.base<- likelihood.ratio.base+x[i,j,k]*log(m[i,j,k]/x[i,j,k])
}
}
}
df<-r*c*t-r-c-t+2
p.Pearson<-pchisq(Pearson.chisq,df=df,lower.tail = FALSE)
likelihood.ratio<- (-2)*likelihood.ratio.base
p.likelihood<-pchisq(likelihood.ratio,df=df,lower.tail = FALSE)
}
if (type=="joint.ind.A"){#for joint independence (A,BC)
for (i in 1:r){
for (j in 1:c){
for (k in 1:t){
m[i,j,k]<-sum(x[i,,])*sum(x[,j,k])/n
Pearson.chisq<-Pearson.chisq+(x[i,j,k]-m[i,j,k])^2/m[i,j,k]
likelihood.ratio.base<- likelihood.ratio.base+x[i,j,k]*log(m[i,j,k]/x[i,j,k])
}
}
}
df<-(r-1)*(c*t-1)
p.Pearson<-pchisq(Pearson.chisq,df=df,lower.tail = FALSE)
likelihood.ratio<- (-2)*likelihood.ratio.base
p.likelihood<-pchisq(likelihood.ratio,df=df,lower.tail = FALSE)
}
if (type=="joint.ind.B"){#for joint independence (B,AC)
for (i in 1:r){
for (j in 1:c){
for (k in 1:t){
m[i,j,k]<-sum(x[,j,])*sum(x[i,,k])/n
Pearson.chisq<-Pearson.chisq+(x[i,j,k]-m[i,j,k])^2/m[i,j,k]
likelihood.ratio.base<- likelihood.ratio.base+x[i,j,k]*log(m[i,j,k]/x[i,j,k])
}
}
}
df<-(c-1)*(r*t-1)
p.Pearson<-pchisq(Pearson.chisq,df=df,lower.tail = FALSE)
likelihood.ratio<- (-2)*likelihood.ratio.base
p.likelihood<-pchisq(likelihood.ratio,df=df,lower.tail = FALSE)
}
if (type=="joint.ind.C"){#for joint independence (C,AB)
for (i in 1:r){
for (j in 1:c){
for (k in 1:t){
m[i,j,k]<-sum(x[,,k])*sum(x[i,j,])/n
Pearson.chisq<-Pearson.chisq+(x[i,j,k]-m[i,j,k])^2/m[i,j,k]
likelihood.ratio.base<- likelihood.ratio.base+x[i,j,k]*log(m[i,j,k]/x[i,j,k])
}
}
}
df<-(t-1)*(r*c-1)
p.Pearson<-pchisq(Pearson.chisq,df=df,lower.tail = FALSE)
likelihood.ratio<- (-2)*likelihood.ratio.base
p.likelihood<-pchisq(likelihood.ratio,df=df,lower.tail = FALSE)
}
if (type=="cond.ind.A"){#for conditional independence (AB,AC)
for (i in 1:r){
for (j in 1:c){
for (k in 1:t){
m[i,j,k]<-sum(x[i,j,])*sum(x[i,,k])/sum(x[i,,])
Pearson.chisq<-Pearson.chisq+(x[i,j,k]-m[i,j,k])^2/m[i,j,k]
likelihood.ratio.base<- likelihood.ratio.base+x[i,j,k]*log(m[i,j,k]/x[i,j,k])
}
}
}
df<-r*(c-1)*(t-1)
p.Pearson<-pchisq(Pearson.chisq,df=df,lower.tail = FALSE)
likelihood.ratio<- (-2)*likelihood.ratio.base
p.likelihood<-pchisq(likelihood.ratio,df=df,lower.tail = FALSE)
}
if (type=="cond.ind.B"){#for conditional independence (BA,BC)
for (i in 1:r){
for (j in 1:c){
for (k in 1:t){
m[i,j,k]<-sum(x[i,j,])*sum(x[,j,k])/sum(x[,j,])
Pearson.chisq<-Pearson.chisq+(x[i,j,k]-m[i,j,k])^2/m[i,j,k]
likelihood.ratio.base<- likelihood.ratio.base+x[i,j,k]*log(m[i,j,k]/x[i,j,k])
}
}
}
df<-c*(r-1)*(t-1)
p.Pearson<-pchisq(Pearson.chisq,df=df,lower.tail = FALSE)
likelihood.ratio<- (-2)*likelihood.ratio.base
p.likelihood<-pchisq(likelihood.ratio,df=df,lower.tail = FALSE)
}
if (type=="cond.ind.C"){#for conditional independence (CA,CB)
for (i in 1:r){
for (j in 1:c){
for (k in 1:t){
m[i,j,k]<-sum(x[i,,k])*sum(x[,j,k])/sum(x[,,k])
Pearson.chisq<-Pearson.chisq+(x[i,j,k]-m[i,j,k])^2/m[i,j,k]
likelihood.ratio.base<- likelihood.ratio.base+x[i,j,k]*log(m[i,j,k]/x[i,j,k])
}
}
}
df<-t*(r-1)*(c-1)
p.Pearson<-pchisq(Pearson.chisq,df=df,lower.tail = FALSE)
likelihood.ratio<- (-2)*likelihood.ratio.base
p.likelihood<-pchisq(likelihood.ratio,df=df,lower.tail = FALSE)
}
return(list(Pearson.chisq=Pearson.chisq,likelihood.ratio=likelihood.ratio,
df=df,p.Pearson=p.Pearson,p.likelihood=p.likelihood))
}
2)三维列联表的相合性检验
对 r x 2 x 2的三维列联表:
- Cochran-Mantel Haenszel 检验(条件相合性检验)
先做分层相合性检验(四格表的Mantel Haenszel Chi2检验),再做条件相合检验 - Breslow-Day 检验(检验分层后各层四格表的相合程度是否相同)
BreslowDaytest<-function(x){
#x is an array including frequencies of r*2*2 contingency table for row A,column B,stratum C.
r<-dim(x)[1]
theta<-NULL;a<-NULL
for (i in 1:r){
theta[i]<-x[i,1,1]*x[i,2,2]/x[i,1,2]/x[i,2,1]
a[i]<-1/x[i,1,1]+1/x[i,1,2]+1/x[i,2,1]+1/x[i,2,2]
}
eta<-log(theta)
etabar<-sum(eta/a)/sum(1/a)
chisq.stat<-sum((eta-etabar)^2/a)
df=r-1
p.chisq<-pchisq(chisq.stat,df=df,lower.tail = FALSE)
return(list(chisq.stat=chisq.stat,df=df,p.chisq=p.chisq))
}
3)三维列联表的相关性检验
- 饱和模型(三个属性全部相关,两两之间、三者之间都有交互作用)
- 两两相关模型 / 齐次相关模型
所有独立性模型都被拒绝后,可以考虑两两相关模型
homogetest<-function(x){
#this function is to test homogeneous association model (AB,AC,BC)
#x is an array including frequencies of r*c*t contingency table for row A,column B,stratum C.
#output includes expected frequencies, Pearson chi-square test and likelihood ratio test
r<-dim(x)[1];c<-dim(x)[2];t<-dim(x)[3]
m0<-m1<-array(1,dim=c(r,c,t)) #initial value
m2<-m3<-array(0,dim=c(r,c,t))
b<-0 #denote number of replications
while (mean(abs(m3-m0))>=10^(-6)){#begin replication
m0<-m1 #update initial value
for (k in 1:t){
for (i in 1:r){
for (j in 1:c){
m1[i,j,k]<- m0[i,j,k]/sum(m0[i,j,])*sum(x[i,j,])
}
}
}
for (j in 1:c){
for (i in 1:r){
for (k in 1:t){
m2[i,j,k]<- m1[i,j,k]/sum(m1[i,,k])*sum(x[i,,k])
}
}
}
for (i in 1:r){
for (j in 1:c){
for (k in 1:t){
m3[i,j,k]<- m2[i,j,k]/sum(m2[,j,k])*sum(x[,j,k])
}
}
}
b<-b+1
m1<-m3
}
m<-m3 #expected frequencies for homogeneous association model (AB,AC,BC)
Pearson.chisq<-likelihood.ratio.base<-0
for (i in 1:r){
for (j in 1:c){
for (k in 1:t){
Pearson.chisq<-Pearson.chisq+(x[i,j,k]-m[i,j,k])^2/m[i,j,k]
likelihood.ratio.base<- likelihood.ratio.base+x[i,j,k]*log(m[i,j,k]/x[i,j,k])
}
}
}
df<-(r-1)*(c-1)*(t-1)
p.Pearson<-pchisq(Pearson.chisq,df=df,lower.tail = FALSE)
likelihood.ratio<- (-2)*likelihood.ratio.base
p.likelihood<-pchisq(likelihood.ratio,df=df,lower.tail = FALSE)
return(list(m=m,replication.no=b,Pearson.chisq=Pearson.chisq,likelihood.ratio=likelihood.ratio,
df=df,p.Pearson=p.Pearson,p.likelihood=p.likelihood))
}
6. Logistic回归模型
1)二分类Logistic回归案例
数据读入和预处理
# 读入原始数据
cuse<- read.table("cuse.txt", header=TRUE)
cuse
summary(cuse)
# 读入赋值和重组变换后的数据
cuseraw<- read.table("cuseraw.txt", header=TRUE)
cuseraw
summary(cuseraw)
attach(cuseraw)
# 频数分布表
Onefactor1<- table(rep(noMore,count),rep(using,count));Onefactor1 # noMore & using
Onefactor2<- table(rep(age,count),rep(using,count));Onefactor2 # age & using
twofactor<- table(rep(noMore,count),rep(age,count),rep(using,count));twofactor # noMore age & using
detach(cuseraw)
# 频数表重组后定义dataframe数据集
cuse.onefactor1<- data.frame(list(wantsMore=c("yes","no"), using=c(219,288),notUsing=c(753,347)));cuse.onefactor1
cuse.onefactor2<- data.frame(list(age=c("<25","25-29","30-39","40-49"),using=c(72,105,237,93),notUsing=c(325,299,375,101)));cuse.onefactor2
cuse.twofactor<- data.frame(list(age=c("<25","<25","25-29","25-29","30-39","30-39","40-49","40-49"),wantsMore=c("yes","no","yes","no","yes","no","yes","no"),
using=c(58,14,68,37,79,158,14,79),notUsing=c(265,60,215,84,230,145,43,58)));cuse.twofactor
Logistic模型建立和分析
① glm(formula, family=binomial(link=logit), data, weights, subset, …);
② the response can be a matrix where the first column is the number of “successes” and the second column is the number of “failures”;
attach(cuse.onefactor1)
## 零模型
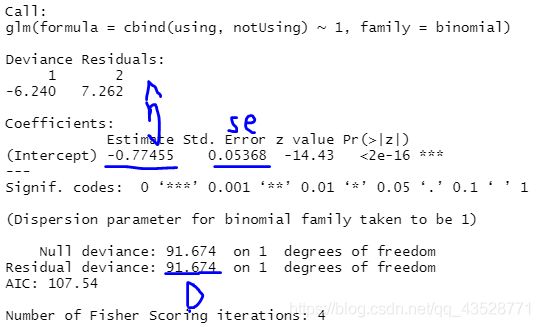
fit.null1<- glm(cbind(using, notUsing) ~ 1, family = binomial)
summary(fit.null1)
## 单因素模型
# R sorts the levels of a factor in alphabetical order, and the first level of a factor is choosen as the reference cell.
noMore <- wantsMore == "no" # 定义示性变量
fit.desire.no<- glm(cbind(using, notUsing) ~ noMore, family = binomial)
summary(fit.desire.no)
# 单因素模型的方差分析表
anova(fit.null1,fit.desire.no)
detach(cuse.onefactor1)
结果分析
- 拟合优度的检验:D大于临界值 → 拟合模型不充分
本例中,零模型拟合不充分(D0),单因素模型拟合充分且就是饱和模型(D1) - 嵌套模型的检验:D(H0) - D(H1)大于临界值 → H1模型增加的效应显著
本例中,效应noMore是显著的
| 零模型 | 单因素模型 |
|---|---|
 |
 |