spark-ml和jpmml-sparkml生成pmml模型过程种遇到的问题
需求:利用pmml(预测模型标记语言)来实现跨平台的机器学习模型部署。
pmml简介:参考链接1
如何将模型生成pmml格式:参考链接3
1、成功的写法:将数据的各种transform和模型全部都放入pipeline中,可以生成pmml。
代码如下:
import org.apache.spark.ml.linalg.Vectors
import org.apache.spark.ml.feature._
import org.apache.spark.sql.SaveMode
import org.apache.spark.sql.SparkSession
import org.apache.spark.sql.functions._
import org.apache.spark.sql.types._
import org.apache.spark.ml.{Pipeline, PipelineModel, PipelineStage}
import org.apache.spark.ml.classification.RandomForestClassifier
import org.apache.spark.ml.classification.RandomForestClassificationModel
import org.jpmml.model.JAXBUtil
import org.jpmml.sparkml.PMMLBuilder
import org.dmg.pmml.PMML
import javax.xml.transform.stream.StreamResult
import java.io.FileOutputStream
import org.apache.spark.ml.linalg.DenseVector
import scala.collection.mutable.ArrayBuffer
object Test extends App{
println("666666")
val spark = SparkSession.builder().master("local").appName("TestPmml").getOrCreate()
val str2Int: Map[String, Double] = Map(
"Iris-setosa" -> 0.0,
"Iris-versicolor" -> 1.0,
"Iris-virginica" -> 2.0
)
var str2double = (x: String) => str2Int(x)
var myFun = udf(str2double)
val data = spark.read.textFile("...\\scalaProgram\\PMML\\iris1.txt").toDF()
.withColumn("splitcol", split(col("value"), ","))
.select(
col("splitcol").getItem(0).as("sepal_length"),
col("splitcol").getItem(1).as("sepal_width"),
col("splitcol").getItem(2).as("petal_length"),
col("splitcol").getItem(3).as("petal_width"),
col("splitcol").getItem(4).as("label")
)
.withColumn("label", myFun(col("label")))
.select(
col("sepal_length").cast(DoubleType),
col("sepal_width").cast(DoubleType),
col("petal_length").cast(DoubleType),
col("petal_width").cast(DoubleType),
col("label").cast(DoubleType)
)
val data1 = data.na.drop()
println("data: " + data1.count().toString)
val schema = data1.schema
println("data1 schema: " + schema)
// merge multi-feature to vector features
val features: Array[String] = Array("sepal_length", "sepal_width", "petal_length", "petal_width")
val assembler: VectorAssembler = new VectorAssembler().setInputCols(features).setOutputCol("features")
val rf: RandomForestClassifier = new RandomForestClassifier()
.setLabelCol("label")
.setFeaturesCol("features")
.setMaxDepth(8)
.setNumTrees(30)
.setSeed(1234)
.setMinInfoGain(0)
.setMinInstancesPerNode(1)
val pipeline = new Pipeline().setStages(Array(assembler,rf))
val pipelineModel = pipeline.fit(newdata1)
println("success fit......")
val pmml = new PMMLBuilder(schema, pipelineModel).build()
val targetFile = "...\\scalaProgram\\PMML\\pipemodel.pmml"
val fis: FileOutputStream = new FileOutputStream(targetFile)
val fout: StreamResult = new StreamResult(fis)
JAXBUtil.marshalPMML(pmml, fout)
println("pmml success......")
}结果:
2、上面代码中VectorAssembler方法就是将多列Double型的数据聚合为一列Vector型的数据。目前因为业务需求,直接给你一列Vector型的数据,然后用模型进行训练并将模型保存为pmml格式。
分析:因为传入模型训练的数据必须是Vector型的,所以上面代码才会利用VectorAssembler将多列属性值合并为一列,而目前已经有了Vector型数据,那就只需要将模型放入Pipeline().setStage()中就行了,试一试
代码:
object Test extends App{
println("666666")
val spark = SparkSession.builder().master("local").appName("TestPmml").getOrCreate()
// convert features string to vector-data
var string2vector = (x: String) => {
var length = x.length()
var a = x.substring(1, length - 1).split(",").map(i => i.toDouble)
Vectors.dense(a)
}
var str2vec = udf(string2vector)
val newdata1 = spark.read.load("...\\scalaProgram\\PMML\\data1.parquet")
val newdata2 = newdata1.withColumn("features", str2vec(col("features")))
println("newdata2: "+newdata2.schema)
val rf: RandomForestClassifier = new RandomForestClassifier()
.setLabelCol("label")
.setFeaturesCol("features")
.setMaxDepth(8)
.setNumTrees(30)
.setSeed(1234)
.setMinInfoGain(0)
.setMinInstancesPerNode(1)
val pipeline = new Pipeline().setStages(Array(rf))
val pipelineModel = pipeline.fit(newdata2)
println("success fit......")
val pmml = new PMMLBuilder(newdata2.schema, pipelineModel).build()
val targetFile = "...\\scalaProgram\\PMML\\pipemodel.pmml"
val fis: FileOutputStream = new FileOutputStream(targetFile)
val fout: StreamResult = new StreamResult(fis)
JAXBUtil.marshalPMML(pmml, fout)
println("pmml success......")
}运行报错:报这个错主要是因为PMMLBuilder中schema里的datatype只支持string,integral,double or boolean,这说明传入pipeline().fit()的原始数据就必须是这些类型。
而我们传入的newdata2数据里features这一列的数据是VectorUDT类型的。根据这个问题,想了一个办法:就是将上面代码中newdata2那一列的数据类型由VectorUDT类型转换为String类型并存成parquet格式,然后从parquet格式中读取出来,那么这一列的数据就是String类型,然后经过VectorAssembler算子,将该算子和模型的算子一起放入pipeline.setStage中,这样貌似也能行,试一试
注意:由于parquet格式是这样的,你写的数据格式是什么类型的,读出来就是相应的类型。
代码:
object TestPmml extends App{
val spark = SparkSession.builder().master("local").appName("TestPmml").getOrCreate()
val str2Int: Map[String, Double] = Map(
"Iris-setosa" -> 0.0,
"Iris-versicolor" -> 1.0,
"Iris-virginica" -> 2.0
)
var str2double = (x: String) => str2Int(x)
var myFun = udf(str2double)
val data = spark.read.textFile("...\\scalaProgram\\PMML\\iris1.txt").toDF()
.withColumn("splitcol", split(col("value"), ","))
.select(
col("splitcol").getItem(0).as("sepal_length"),
col("splitcol").getItem(1).as("sepal_width"),
col("splitcol").getItem(2).as("petal_length"),
col("splitcol").getItem(3).as("petal_width"),
col("splitcol").getItem(4).as("label")
)
.withColumn("label", myFun(col("label")))
.select(
col("sepal_length").cast(DoubleType),
col("sepal_width").cast(DoubleType),
col("petal_length").cast(DoubleType),
col("petal_width").cast(DoubleType),
col("label").cast(DoubleType)
)
val data1 = data.na.drop()
println("data: " + data1.count().toString)
val schema = data1.schema
println("data1 schema: " + schema)
val features: Array[String] = Array("sepal_length", "sepal_width", "petal_length", "petal_width")
// // merge multi-feature to vector features
val assembler: VectorAssembler = new VectorAssembler().setInputCols(features).setOutputCol("features")
val data2 = assembler.transform(data1)
println("data2 schema: " + data2.schema)
println("assembler transform class: "+assembler.getClass )
// convert features vector-data to string
val convertFunction = (x: DenseVector) => {
x.toString
}
val convertUDF = udf(convertFunction)
val newdata = data2.withColumn("features", convertUDF(col("features")))
newdata.write.mode(SaveMode.Overwrite).format("parquet").save("...\\scalaProgram\\PMML\\data1.parquet")
val newdata1 = spark.read.load("...\\scalaProgram\\PMML\\data1.parquet")
val assembler1: VectorAssembler = new VectorAssembler().setInputCols(features).setOutputCol("features")
val rf = new RandomForestClassifier()
.setLabelCol("label")
.setFeaturesCol("features")
.setMaxDepth(8)
.setNumTrees(30)
.setSeed(1234)
.setMinInfoGain(0)
.setMinInstancesPerNode(1)
val pipeline = new Pipeline().setStages(Array(mytransformer, rf))
//
val pipelineModel = pipeline.fit(newdata1)
// val pre = pipelineModel.transform(data)
//val prediction = pre.select("prediction")
// import org.apache.spark.ml.evaluation.MulticlassClassificationEvaluator
// val evaluator = new MulticlassClassificationEvaluator()
// .setLabelCol("label").setMetricName("accuracy").setPredictionCol("prediction")
// val acc = evaluator.evaluate(pre)
// print("acc "+acc)
val pmml = new PMMLBuilder(newdata1.schema, pipelineModel).build()
val targetFile = "...\\scalaProgram\\PMML\\pipemodel.pmml"
val fis: FileOutputStream = new FileOutputStream(targetFile)
val fout: StreamResult = new StreamResult(fis)
JAXBUtil.marshalPMML(pmml, fout)
}运行报错:fit这一步就直接报错了,是说原始数据经过transformer算子后的features列的数据类型是String(传入模型训练的数据必须是Vector型的),不是vector类型
这样看来,原本是String类型的输入数据,经过VectorAssembler算子后,数据并没有转换为Vector型,这说明VectorAssembler算子只是将非Vector数据变为Vector数据,但是不会改变数据的具体类型,所以这种方法不行。那么可以自定义一个transformer算子(官方支持开发),这个算子将原来是String类型的数据变为Vector型,这样的话就既满足了之前原始数据是string类型,经过transformer算子变换后,输入到模型的数据是Vector型,看起来有些道理,试一试
自定义transformer算子可以参考:(不能全部参考,因为里面没有HasInputCol(s)和HasOutputCol(s),下面会说到)
https://zhuanlan.zhihu.com/p/27687260
https://my.oschina.net/weekn/blog/1975783
代码:
自定义transformer算子:Mytransformer.scala,注意:package org.apache.spark.ml.feature一定得加上,不然会出现如下问题
package org.apache.spark.ml.feature
import java.util.NoSuchElementException
import scala.collection.mutable
import scala.language.existentials
import org.apache.spark.SparkException
import org.apache.spark.annotation.Since
import org.apache.spark.ml.Transformer
import org.apache.spark.ml.attribute.{Attribute, AttributeGroup, NumericAttribute, UnresolvedAttribute}
import org.apache.spark.ml.linalg.{Vector, VectorUDT, Vectors}
import org.apache.spark.ml.param.{Param, ParamMap, ParamValidators}
import org.apache.spark.ml.param.shared._
import org.apache.spark.ml.util._
import org.apache.spark.mllib.linalg.Vectors
import org.apache.spark.sql.{DataFrame, Dataset, Row}
import org.apache.spark.sql.functions._
import org.apache.spark.sql.types._
import org.apache.spark.annotation.Since
class Mytransformer(override val uid: String) extends Transformer with HasInputCols with HasOutputCols with DefaultParamsWritable {
final val inputCol= new Param[String](this, "inputCol", "The input column")
final val outputCol = new Param[String](this, "outputCol", "The output column")
@Since("1.4.0")
def setInputCol(value: String): this.type = set(inputCol, value)
@Since("1.4.0")
def setOutputCol(value: String): this.type = set(outputCol, value)
def this() = this(Identifiable.randomUID("Mytransformer "))
@Since("1.4.1")
override def copy(extra: ParamMap): Mytransformer = {
defaultCopy(extra)
}
@Since("1.4.0")
override def transformSchema(schema: StructType): StructType = {
val idx = schema.fieldIndex($(inputCol))
val field = schema.fields(idx)
// if (field.dataType != DoubleType) {
// throw new Exception(s"Input type ${field.dataType} did not match input type DoubleType")
// }
schema.add(StructField($(outputCol), new VectorUDT, false))
}
@Since("2.0.0")
override def transform(df: Dataset[_]):DataFrame = {
var string2vector = (x: String) => {
var length = x.length()
var a = x.substring(1, length - 1).split(",").map(i => i.toDouble)
org.apache.spark.ml.linalg.Vectors.dense(a)
}
var str2vec = udf(string2vector)
df.withColumn($(outputCol), str2vec(col($(inputCol))))
}
// @Since("1.6.0")
// override def load(path:String):Mytransformer = super.load(path)
}
......
import org.apache.spark.ml.feature.Mytransformer
object TestPmml extends App{
val spark = SparkSession.builder().master("local").appName("TestPmml").getOrCreate()
val str2Int: Map[String, Double] = Map(
"Iris-setosa" -> 0.0,
"Iris-versicolor" -> 1.0,
"Iris-virginica" -> 2.0
)
var str2double = (x: String) => str2Int(x)
var myFun = udf(str2double)
val data = spark.read.textFile("...\\scalaProgram\\PMML\\iris1.txt").toDF()
.withColumn("splitcol", split(col("value"), ","))
.select(
col("splitcol").getItem(0).as("sepal_length"),
col("splitcol").getItem(1).as("sepal_width"),
col("splitcol").getItem(2).as("petal_length"),
col("splitcol").getItem(3).as("petal_width"),
col("splitcol").getItem(4).as("label")
)
.withColumn("label", myFun(col("label")))
.select(
col("sepal_length").cast(DoubleType),
col("sepal_width").cast(DoubleType),
col("petal_length").cast(DoubleType),
col("petal_width").cast(DoubleType),
col("label").cast(DoubleType)
)
val data1 = data.na.drop()
println("data: " + data1.count().toString)
val schema = data1.schema
println("data1 schema: " + schema)
val features: Array[String] = Array("sepal_length", "sepal_width", "petal_length", "petal_width")
// merge multi-feature to vector features
val assembler: VectorAssembler = new VectorAssembler().setInputCols(features).setOutputCol("features")
val data2 = assembler.transform(data1)
// convert features vector-data to string
val convertFunction = (x: DenseVector) => {
x.toString
}
val convertUDF = udf(convertFunction)
val newdata = data2.withColumn("features", convertUDF(col("features")))
newdata.write.mode(SaveMode.Overwrite).format("parquet").save("...\\scalaProgram\\PMML\\data1.parquet")
// convert features string to vector-data
var string2vector = (x: String) => {
var length = x.length()
var a = x.substring(1, length - 1).split(",").map(i => i.toDouble)
Vectors.dense(a)
}
var str2vec = udf(string2vector)
val newdata1 = spark.read.load("...\\scalaProgram\\PMML\\data1.parquet")
println("newdata1: " + newdata1.schema)
val mytransformer = new Mytransformer().setInputCol("features").setOutputCol("features")
val rf = new RandomForestClassifier()
.setLabelCol("label")
.setFeaturesCol("features")
.setMaxDepth(8)
.setNumTrees(30)
.setSeed(1234)
.setMinInfoGain(0)
.setMinInstancesPerNode(1)
val pipeline = new Pipeline().setStages(Array(mytransformer, rf))
val pipelineModel = pipeline.fit(newdata1)
val pmml = new PMMLBuilder(newdata1.schema, pipelineModel).build()
val targetFile = "...\\scalaProgram\\PMML\\pipemodel.pmml"
val fis: FileOutputStream = new FileOutputStream(targetFile)
val fout: StreamResult = new StreamResult(fis)
JAXBUtil.marshalPMML(pmml, fout)
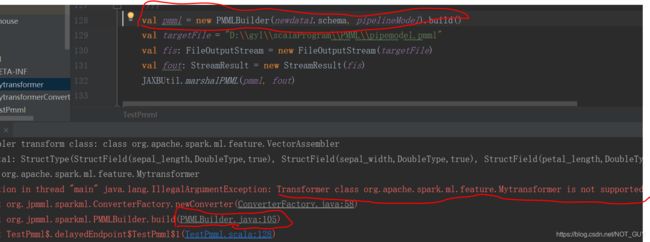
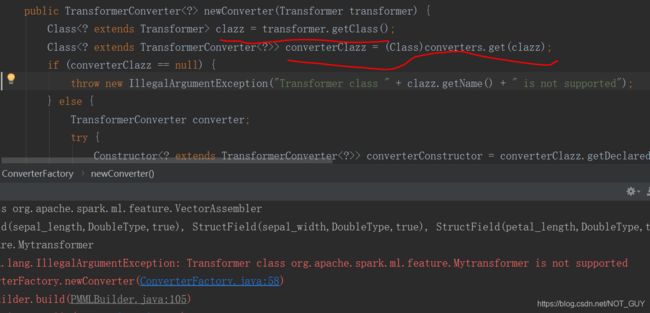
}运行报错:这是说jpmml-spark库中不支持自定义的transformer算子
进一步查看,发现converters变量是一个map变量,它里面的key是org.apache.spark.ml.feature里的transformer算子(github上jpmml-spark中features里写明了),value是jpmml-spark中feature里对应算子的transformerConverter函数,而我们自定义的算子Converter函数并没有相应的在里面,所以报错了。
![]()
进一步分析,假如把我们自定义的transformerConverter放入jpmml-spark库的feature中,然后生成自己的jar包,这样是不是就行了呢,试了试,发现还是不行,首先在生成jar时,会报错:unable to guarantee security of recursive delete。试了一下强行生成jar包,然后放入自己的工程中,运行发现还是报错:Transformer class org.apache.spark.ml.feature.Mytransformer is not supporter。
最后,还是选择直接问问jpmml-spark库的维护人员吧,更新中。。。。。。
——————————————————————————————————————————————————————
问题终于解决了,再更新一拨吧。。。。。。
在github上问了一下jpmml-spark库的维护人员,给我的回复是https://github.com/jpmml/jpmml-sparkml/issues/72(竟然有人在一个月前问过了(手动苦笑,不过这里得出一个经验,以后有问题就多看看这个开源库旁边的issue,我还是太菜了),维护人员效率很高,回答问题很快(问题发出去后,22s就给了回复),感谢大佬的帮助。。。。。。
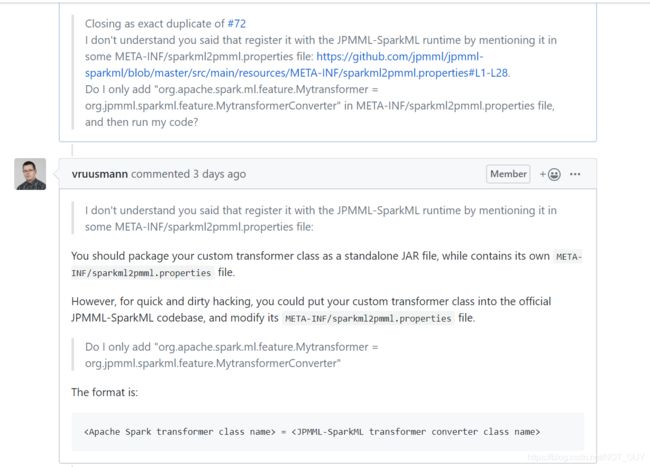
可以清楚看到解决办法很简单:
1、用Scala写你自定义的transformer类;2、用java或scala写相应的transformer-to-pmml-converter类(这个其实就是一个converter类,可以仿照org.jpmml.sparkml.feature里的写);3、将前面写的两个类放在自己项目路径里,然后在META-INF/sparkml2pmml.properties提及它,当程序运行时,JPMML-SparkML就会register。(在META-INF里新建一个sparkml2pmml.properties,这个文件在ConverterFactory.class中会被解析成一个Map并赋值给converters变量,然后在这个文件里写一条语句,具体的写法可以参考官方写的,很容易:
org.apache.spark.ml.feature.Mytransformer = org.jpmml.sparkml.feature.MytransformerConverter由于本人太垃圾了,导致在第3条里的register。。。困惑了很久,不知道怎么弄,然后又问了一下维护人员,他给出的答复是
图中他给了两种方法,但最后我并没有这样做(其实第二种方法在问之前我试过,直接修改源码生成jar包,但是生成jar包的过程中会报18个recursive错误和不安全警告,毕竟是dirty hacking,哈哈哈。。。。。。),而是在网上看到别人自定义自己的xgboost模型的做法是如何放置的这些文件才明白register.......这句话的意思:(就是上面的解释)https://blog.csdn.net/baifanwudi/article/details/86607906。
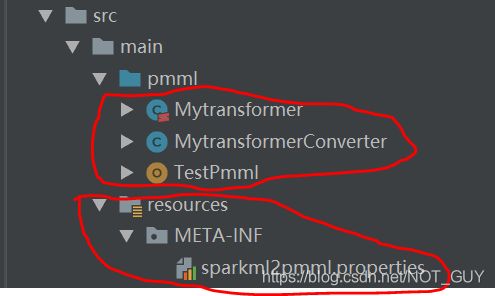
三种文件的路径结构:
自定义的transformer类:Mytransformer.scala
package org.apache.spark.ml.feature
import java.util.NoSuchElementException
import scala.collection.mutable
import scala.language.existentials
import org.apache.spark.SparkException
import org.apache.spark.annotation.Since
import org.apache.spark.ml.Transformer
import org.apache.spark.ml.attribute.{Attribute, AttributeGroup, NumericAttribute, UnresolvedAttribute}
import org.apache.spark.ml.linalg.{Vector, VectorUDT, Vectors}
import org.apache.spark.ml.param.{Param, ParamMap, ParamValidators}
import org.apache.spark.ml.param.shared._
import org.apache.spark.ml.util._
import org.apache.spark.mllib.linalg.Vectors
import org.apache.spark.sql.{DataFrame, Dataset, Row}
import org.apache.spark.sql.functions._
import org.apache.spark.sql.types._
//import org.apache.spark.annotation.Since
// 一定要有HasOutputCol,jpmml-spark里FeatureConverter.class的registerFeatures函数会用到,不然会报错; 到底是用HasInputCols还是HasInputCol,取决于你传入的参数是Array类型还是基本类型。
class Mytransformer(override val uid: String) extends Transformer with HasInputCols with HasOutputCol{
// 可不写
// final val inputCol= new Param[String](this, "inputCol", "The input column")
// final val outputCol = new Param[String](this, "outputCol", "The output column")
// 注意HasInputCols对应的是inputCols, 值是Array型, HasInputCol对应的是inputCol, 值是基本数据类型(不是Array型), HasOutputCol同理
def setInputCol(value: Array[String]): this.type = set(inputCols, value)
def setOutputCol(value: String): this.type = set(outputCol, value)
def this() = this(Identifiable.randomUID("Mytransformer "))
override def copy(extra: ParamMap): Mytransformer = {
defaultCopy(extra)
}
@Since("1.4.0")
override def transformSchema(schema: StructType): StructType = {
// 对输入特征的数据类型进行判断
// val inputColNames = $(inputCols)
// val outputColName = $(outputCol)
// val incorrectColumns = inputColNames.flatMap { name =>
// schema(name).dataType match {
// case _: NumericType | BooleanType => None
// case t if t.isInstanceOf[VectorUDT] => None
// case other => Some(s"Data type ${other.catalogString} of column $name is not supported.")
// }
//}
schema.add(StructField($(outputCol), new VectorUDT, false))
}
override def transform(df: Dataset[_]):DataFrame = {
// 这个transform函数只是对df中某一列数据进行处理
var string2vector = (x: String) => {
var length = x.length()
var a = x.substring(1, length - 1).split(",").map(i => i.toDouble)
org.apache.spark.ml.linalg.Vectors.dense(a)
}
var str2vec = udf(string2vector)
// str2vec函数中传入你要处理的df中的列名
df.withColumn($(outputCol), str2vec(col("features")))
}
}
transformerConverter类:MytransformerConverter.java
// 这句虽然有红线,但是不能去掉,否则报错
package org.jpmml.sparkml.feature;
import java.util.ArrayList;
import java.util.List;
import org.apache.spark.ml.feature.Mytransformer;
import org.jpmml.converter.Feature;
import org.jpmml.sparkml.FeatureConverter;
import org.jpmml.sparkml.SparkMLEncoder;
// 参考jpmml-spark中feature里的VectorAssemblerConverter.java写
public class MytransformerConverter extends FeatureConverter {
public MytransformerConverter(Mytransformer transformer){
super(transformer);
}
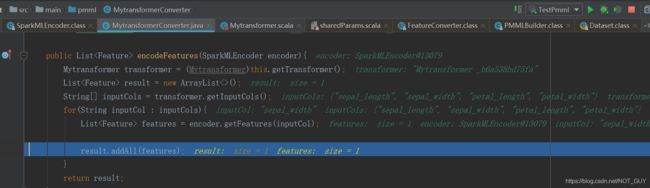
public List encodeFeatures(SparkMLEncoder encoder){
Mytransformer transformer = (Mytransformer)this.getTransformer();
List result = new ArrayList<>();
String[] inputCols = transformer.getInputCols();
for(String inputCol : inputCols){
List features = encoder.getFeatures(inputCol);
result.addAll(features);
}
return result;
}
} sparkml2pmml.properties文件配置:这里,本人是把库里的全部复制过来,然后添加了自己的定义的——如下代码第一句(对应上面的mentioning it in some META-INF/sparkml2pmml.properties)
# Features
org.apache.spark.ml.feature.Mytransformer = org.jpmml.sparkml.feature.MytransformerConverter
org.apache.spark.ml.feature.Binarizer = org.jpmml.sparkml.feature.BinarizerConverter
org.apache.spark.ml.feature.Bucketizer = org.jpmml.sparkml.feature.BucketizerConverter
org.apache.spark.ml.feature.ChiSqSelectorModel = org.jpmml.sparkml.feature.ChiSqSelectorModelConverter
org.apache.spark.ml.feature.ColumnPruner = org.jpmml.sparkml.feature.ColumnPrunerConverter
org.apache.spark.ml.feature.CountVectorizerModel = org.jpmml.sparkml.feature.CountVectorizerModelConverter
org.apache.spark.ml.feature.IDFModel = org.jpmml.sparkml.feature.IDFModelConverter
org.apache.spark.ml.feature.ImputerModel = org.jpmml.sparkml.feature.ImputerModelConverter
org.apache.spark.ml.feature.IndexToString = org.jpmml.sparkml.feature.IndexToStringConverter
org.apache.spark.ml.feature.Interaction = org.jpmml.sparkml.feature.InteractionConverter
org.apache.spark.ml.feature.MaxAbsScalerModel = org.jpmml.sparkml.feature.MaxAbsScalerModelConverter
org.apache.spark.ml.feature.MinMaxScalerModel = org.jpmml.sparkml.feature.MinMaxScalerModelConverter
org.apache.spark.ml.feature.NGram = org.jpmml.sparkml.feature.NGramConverter
org.apache.spark.ml.feature.OneHotEncoder = org.jpmml.sparkml.feature.OneHotEncoderConverter
org.apache.spark.ml.feature.OneHotEncoderModel = org.jpmml.sparkml.feature.OneHotEncoderModelConverter
org.apache.spark.ml.feature.PCAModel = org.jpmml.sparkml.feature.PCAModelConverter
org.apache.spark.ml.feature.RegexTokenizer = org.jpmml.sparkml.feature.RegexTokenizerConverter
org.apache.spark.ml.feature.RFormulaModel = org.jpmml.sparkml.feature.RFormulaModelConverter
org.apache.spark.ml.feature.SQLTransformer = org.jpmml.sparkml.feature.SQLTransformerConverter
org.apache.spark.ml.feature.StandardScalerModel = org.jpmml.sparkml.feature.StandardScalerModelConverter
org.apache.spark.ml.feature.StringIndexerModel = org.jpmml.sparkml.feature.StringIndexerModelConverter
org.apache.spark.ml.feature.StopWordsRemover = org.jpmml.sparkml.feature.StopWordsRemoverConverter
org.apache.spark.ml.feature.Tokenizer = org.jpmml.sparkml.feature.TokenizerConverter
org.apache.spark.ml.feature.VectorAssembler = org.jpmml.sparkml.feature.VectorAssemblerConverter
org.apache.spark.ml.feature.VectorAttributeRewriter = org.jpmml.sparkml.feature.VectorAttributeRewriterConverter
org.apache.spark.ml.feature.VectorIndexerModel = org.jpmml.sparkml.feature.VectorIndexerModelConverter
org.apache.spark.ml.feature.VectorSizeHint = org.jpmml.sparkml.feature.VectorSizeHintConverter
org.apache.spark.ml.feature.VectorSlicer = org.jpmml.sparkml.feature.VectorSlicerConverter
# Prediction models
org.apache.spark.ml.classification.DecisionTreeClassificationModel = org.jpmml.sparkml.model.DecisionTreeClassificationModelConverter
org.apache.spark.ml.classification.GBTClassificationModel = org.jpmml.sparkml.model.GBTClassificationModelConverter
org.apache.spark.ml.classification.LinearSVCModel = org.jpmml.sparkml.model.LinearSVCModelConverter
org.apache.spark.ml.classification.LogisticRegressionModel = org.jpmml.sparkml.model.LogisticRegressionModelConverter
org.apache.spark.ml.classification.MultilayerPerceptronClassificationModel = org.jpmml.sparkml.model.MultilayerPerceptronClassificationModelConverter
org.apache.spark.ml.classification.NaiveBayesModel = org.jpmml.sparkml.model.NaiveBayesModelConverter
org.apache.spark.ml.classification.RandomForestClassificationModel = org.jpmml.sparkml.model.RandomForestClassificationModelConverter
org.apache.spark.ml.clustering.KMeansModel = org.jpmml.sparkml.model.KMeansModelConverter
org.apache.spark.ml.regression.DecisionTreeRegressionModel = org.jpmml.sparkml.model.DecisionTreeRegressionModelConverter
org.apache.spark.ml.regression.GBTRegressionModel = org.jpmml.sparkml.model.GBTRegressionModelConverter
org.apache.spark.ml.regression.GeneralizedLinearRegressionModel = org.jpmml.sparkml.model.GeneralizedLinearRegressionModelConverter
org.apache.spark.ml.regression.LinearRegressionModel = org.jpmml.sparkml.model.LinearRegressionModelConverter
org.apache.spark.ml.regression.RandomForestRegressionModel = org.jpmml.sparkml.model.RandomForestRegressionModelConverter
其实,在最后成功之前,还碰到一个问题:Expected 4 feature(s), got 1 feature(s).
通过debug发现,pipelineModel里的numFeatures值为4,所以才导致这个bug的。
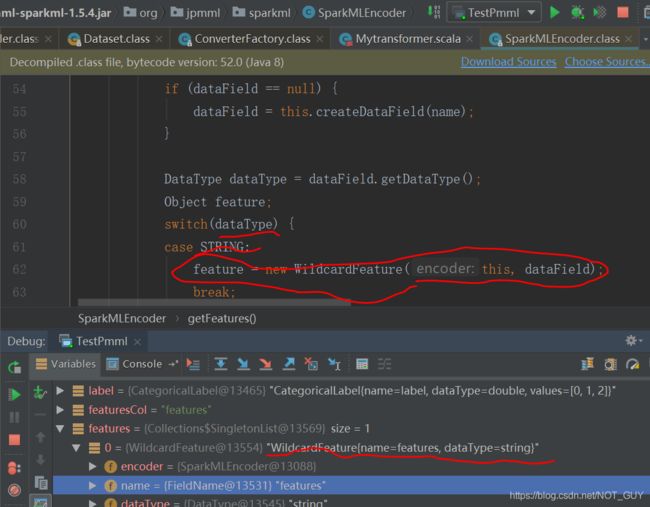
然后,debug程序发现PMMLBuilder.class里运行至第87行的
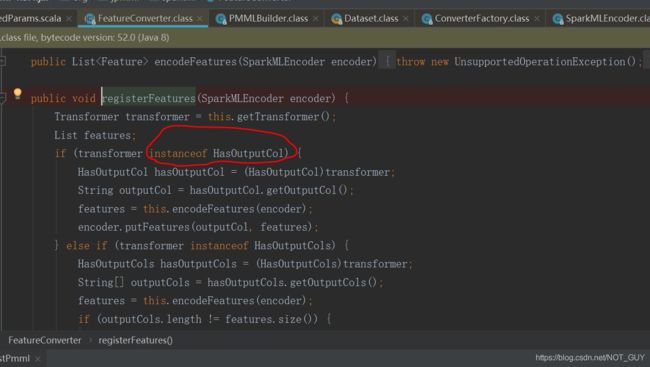
featureConverter.registerFeatures(encoder);,进入registerFeatures函数体后,因为transformer没有HasOutputCol(这就是上面为啥说不能全部参考别人的,要根据实际的来操作),导致encoder里面就没有features,导致最后SparkMLEncoder.class里的getFeatures(String column)没有执行MytransformerConverter.java里的encoderFeatures函数(关键原因,这是通过debug最前面那个成功的代码才发现的)。
至此,利用scala+spark+JPmml-Spark库将模型转换成pmml格式就成功了。这个过程中,收益很多,最后还是感谢JPmml-Spark库的维护人员以及领导的帮助。
代码见github:https://github.com/GuoYL36/myTransformer_Spark
参考链接:
1、https://www.cnblogs.com/pinard/p/9220199.html
2、https://github.com/jpmml/jpmml-sparkml
3、https://blog.csdn.net/baifanwudi/article/details/83892730
4、https://my.oschina.net/weekn/blog/1975845