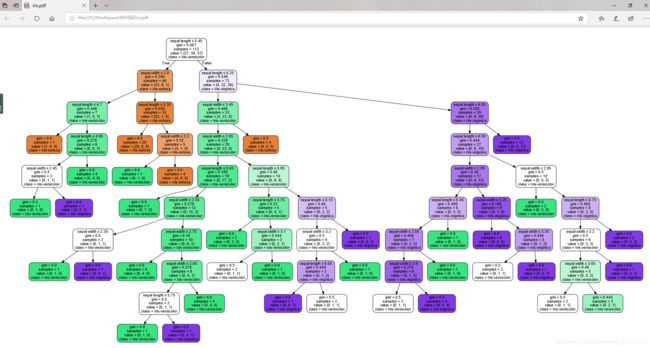
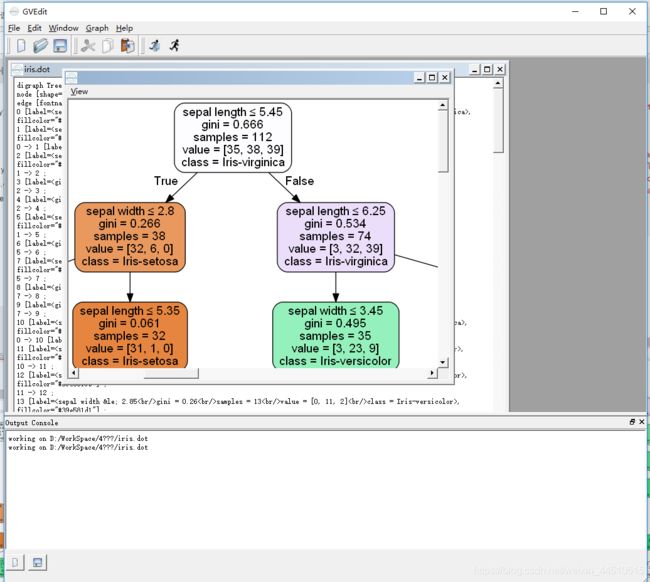
决策树如何可视化
如果不能将一棵决策树可视化,我觉的很难学好决策树这一部分
安装好Graphviz
为什么要安装呢
因为要使用sklearn自带的 export_graphviz
http://www.graphviz.org/
pip install pydotplus
测试一下
# -*- coding:utf-8 -*-
# time :2019/4/18 13:33
# author: 毛利
from sklearn.model_selection import train_test_split
import pandas as pd
from sklearn.tree import DecisionTreeClassifier
from sklearn import datasets
from sklearn import tree
from sklearn.metrics import accuracy_score
import pydotplus
iris = datasets.load_iris()
iris_feature = '花萼长度', '花萼宽度', '花瓣长度', '花瓣宽度'
iris_feature_E = 'sepal length', 'sepal width', 'petal length', 'petal width'
iris_class = 'Iris-setosa', 'Iris-versicolor', 'Iris-virginica'
x = pd.DataFrame(iris['data'])[[0,1]]
y = pd.Series(iris[ 'target'])
x_train,x_test,y_train,y_test = train_test_split(x,y)
model = DecisionTreeClassifier()
model.fit(x_train,y_train)
y_train_pred = model.predict(x_train)
print('训练集正确率:', accuracy_score(y_train, y_train_pred))
data = tree.export_graphviz(model, out_file='iris.dot', feature_names= iris_feature_E[0:2], class_names=iris_class,
filled=True, rounded=True, special_characters=True)
graph = pydotplus.graph_from_dot_data(data)
graph.write_pdf('iris.pdf')
with open('iris.png', 'wb') as f:
f.write(graph.create_png())、
import numpy as np
from sklearn.tree import DecisionTreeClassifier
import pydotplus
from sklearn import tree
X = np.array([[2, 2],
[2, 1],
[2, 3],
[1, 2],
[1, 1],
[3, 3]])
y = np.array([0, 1, 1, 1, 0, 1])
plt.style.use('fivethirtyeight')
plt.rcParams['font.size'] = 18
plt.figure(figsize=(8, 8))
# Plot each point as the label
for x1, x2, label in zip(X[:, 0], X[:, 1], y):
plt.text(x1, x2, str(label), fontsize=40, color='g',
ha='center', va='center')
plt.grid(None)
plt.xlim((0, 3.5))
plt.ylim((0, 3.5))
plt.xlabel('x1', size=20)
plt.ylabel('x2', size=20)
plt.title('Data', size=24)
# plt.show()
dec_tree = DecisionTreeClassifier()
print(dec_tree)
dec_tree.fit(X, y)
print(dec_tree.score(X,y))
# Export as dot
dot_data = tree.export_graphviz(dec_tree, out_file=None,
feature_names=['x1', 'x2'],
class_names=['0', '1'],
filled=True, rounded=True,
special_characters=True)
graph = pydotplus.graph_from_dot_data(dot_data)
with open('1.png', 'wb') as f:
f.write(graph.create_png())
def export_graphviz(decision_tree, out_file=SENTINEL, max_depth=None,
feature_names=None, class_names=None, label='all',
filled=False, leaves_parallel=False, impurity=True,
node_ids=False, proportion=False, rotate=False,
rounded=False, special_characters=False, precision=3):
"""Export a decision tree in DOT format.
This function generates a GraphViz representation of the decision tree,
which is then written into `out_file`. Once exported, graphical renderings
can be generated using, for example::
$ dot -Tps tree.dot -o tree.ps (PostScript format)
$ dot -Tpng tree.dot -o tree.png (PNG format)
The sample counts that are shown are weighted with any sample_weights that
might be present.
Read more in the :ref:`User Guide `.
Parameters
----------
decision_tree : decision tree classifier
The decision tree to be exported to GraphViz.
out_file : file object or string, optional (default='tree.dot')
Handle or name of the output file. If ``None``, the result is
returned as a string. This will the default from version 0.20.
max_depth : int, optional (default=None)
The maximum depth of the representation. If None, the tree is fully
generated.
feature_names : list of strings, optional (default=None)
Names of each of the features.
class_names : list of strings, bool or None, optional (default=None)
Names of each of the target classes in ascending numerical order.
Only relevant for classification and not supported for multi-output.
If ``True``, shows a symbolic representation of the class name.
label : {'all', 'root', 'none'}, optional (default='all')
Whether to show informative labels for impurity, etc.
Options include 'all' to show at every node, 'root' to show only at
the top root node, or 'none' to not show at any node.
filled : bool, optional (default=False)
When set to ``True``, paint nodes to indicate majority class for
classification, extremity of values for regression, or purity of node
for multi-output.
leaves_parallel : bool, optional (default=False)
When set to ``True``, draw all leaf nodes at the bottom of the tree.
impurity : bool, optional (default=True)
When set to ``True``, show the impurity at each node.
node_ids : bool, optional (default=False)
When set to ``True``, show the ID number on each node.
proportion : bool, optional (default=False)
When set to ``True``, change the display of 'values' and/or 'samples'
to be proportions and percentages respectively.
rotate : bool, optional (default=False)
When set to ``True``, orient tree left to right rather than top-down.
rounded : bool, optional (default=False)
When set to ``True``, draw node boxes with rounded corners and use
Helvetica fonts instead of Times-Roman.
special_characters : bool, optional (default=False)
When set to ``False``, ignore special characters for PostScript
compatibility.
precision : int, optional (default=3)
Number of digits of precision for floating point in the values of
impurity, threshold and value attributes of each node.
Returns
-------
dot_data : string
String representation of the input tree in GraphViz dot format.
Only returned if ``out_file`` is None.
.. versionadded:: 0.18