R语言排序分析(PCA、CA、PCoA、NMDS、RDA)
文章目录
-
- ****排序 Ordination****
- 非约束性排序
-
- PCA
- CA
- PCoA
- NMDS
- **RDA约束性排序**
-
- ggplot可视化
- ggvegan可视化
- rda.plot()函数
排序 Ordination
按照有无解释变量,可分为非约束性排序(unconstrained ordination)与约束性排序(constrained ordination)。
非约束性排序
- PCA 线性模型,基于原始矩阵
- CA 单峰模型,基于原始矩阵
- PCoA 基于原始距离矩阵
- NMDS 基于秩和距离矩阵
- Clustering
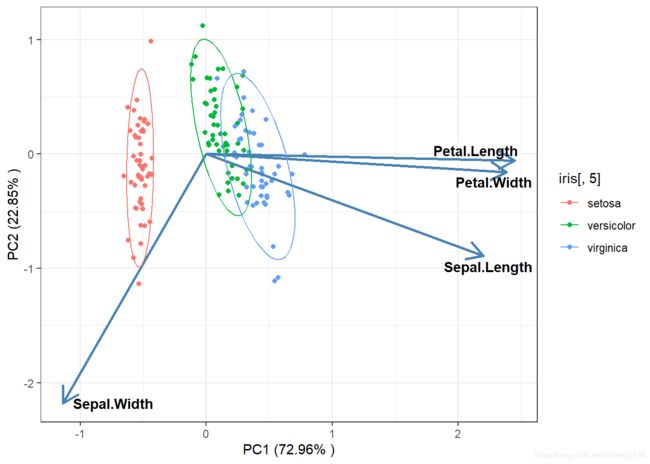
PCA
PCA计算的是欧氏距离。
library(vegan)
library(tidyverse)
library(ggpubr)
#l对iris做PCA
model_pca<-rda(iris[,-5],scale=T,scaling=1)
#提取特征值与方差解释度%
summary(model_pca)$cont$importance #特征值与方差解释度
## Importance of components:
## PC1 PC2 PC3 PC4
## Eigenvalue 2.9185 0.9140 0.14676 0.020715
## Proportion Explained 0.7296 0.2285 0.03669 0.005179
## Cumulative Proportion 0.7296 0.9581 0.99482 1.000000
#载荷矩阵loading
head(model_pca$CA$v)
## PC1 PC2 PC3 PC4
## Sepal.Length 0.5210659 -0.37741762 0.7195664 0.2612863
## Sepal.Width -0.2693474 -0.92329566 -0.2443818 -0.1235096
## Petal.Length 0.5804131 -0.02449161 -0.1421264 -0.8014492
## Petal.Width 0.5648565 -0.06694199 -0.6342727 0.5235971
#site scores
sites_score<-scores(model_pca,display = "sites")%>%
as.data.frame()%>%
rownames_to_column()%>%
as_tibble()
sites_score
## # A tibble: 150 x 3
## rowname PC1 PC2
##
## 1 sit1 -0.535 -0.203
## 2 sit2 -0.491 0.284
## 3 sit3 -0.558 0.144
## 4 sit4 -0.543 0.252
## 5 sit5 -0.564 -0.273
## 6 sit6 -0.490 -0.628
## 7 sit7 -0.577 -0.0201
## 8 sit8 -0.527 -0.0942
## 9 sit9 -0.551 0.471
## 10 sit10 -0.516 0.198
## # ... with 140 more rows
#specoes scores
species_score<-scores(model_pca,display = "species")%>%
as.data.frame()%>%
rownames_to_column()%>%
as_tibble()
species_score
## # A tibble: 4 x 3
## rowname PC1 PC2
##
## 1 Sepal.Length 2.20 -0.891
## 2 Sepal.Width -1.14 -2.18
## 3 Petal.Length 2.45 -0.0578
## 4 Petal.Width 2.38 -0.158
#可视化
ggplot()+
geom_point(data=sites_score,aes(x=PC1,y=PC2,color=iris[,5]))+
labs(x=paste("PC1 (",round(summary(model_pca)$cont$importance [2,1]*100,2)
,"% )",sep=""),
y=paste("PC2 (",round(summary(model_pca)$cont$importance [2,2]*100,2)
,"% )",sep=""))+
geom_segment(data=species_score,aes(x=0,y=0,xend=PC1,yend=PC2),size = 1,
arrow = arrow(length = unit(0.2, "inches")),color="steelblue")+
ggrepel::geom_text_repel(data=species_score,nudge_x = 0.4,
fontface="bold", color="black",
aes(x=PC1,y=PC2,label=rowname))+
stat_ellipse(type="norm",level= 0.9,
data=sites_score,aes(x=PC1,y=PC2,color=iris[,5]))+
theme_bw()
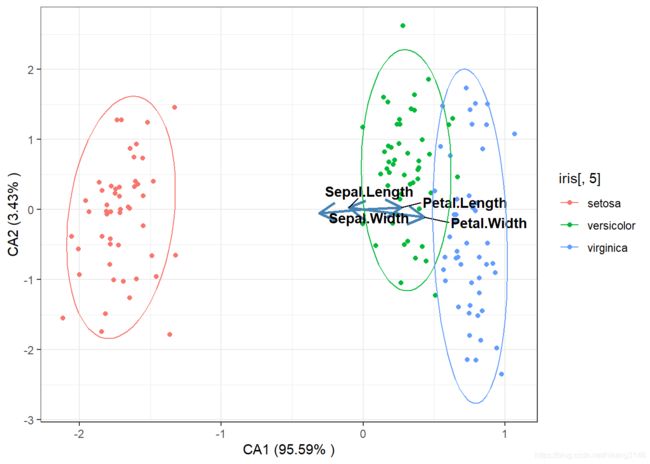
CA
CA计算用的是卡方距离。
#l对iris做PCA
model_ca<-cca(iris[,-5],scale=T,scaling=1)
#提取特征值与方差解释度%
summary(model_ca)$cont$importance #特征值与方差解释度
## Importance of components:
## CA1 CA2 CA3
## Eigenvalue 0.06129 0.00220 0.0006302
## Proportion Explained 0.95585 0.03432 0.0098288
## Cumulative Proportion 0.95585 0.99017 1.0000000
#载荷矩阵loading
head(model_ca$CA$v)
## CA1 CA2 CA3
## Sepal.Length -0.4118927 0.6554888 0.8787892
## Sepal.Width -1.2425352 -1.0612937 -0.9286927
## Petal.Length 1.0958023 0.5920802 -1.0659541
## Petal.Width 1.7406720 -2.3434255 1.4258929
#site scores
sites_score<-scores(model_ca,display = "sites")%>%
as.data.frame()%>%
rownames_to_column()%>%
as_tibble()
sites_score
## # A tibble: 150 x 3
## rowname CA1 CA2
##
## 1 sit1 -1.81 -0.0236
## 2 sit2 -1.64 0.871
## 3 sit3 -1.78 -0.0325
## 4 sit4 -1.61 0.328
## 5 sit5 -1.84 -0.382
## 6 sit6 -1.60 -0.992
## 7 sit7 -1.69 -1.03
## 8 sit8 -1.72 0.187
## 9 sit9 -1.60 0.399
## 10 sit10 -1.71 1.28
## # ... with 140 more rows
#species scores
species_score<-scores(model_ca,display = "species")%>%
as.data.frame()%>%
rownames_to_column()%>%
as_tibble()
species_score
## # A tibble: 4 x 3
## rowname CA1 CA2
##
## 1 Sepal.Length -0.102 0.0307
## 2 Sepal.Width -0.308 -0.0498
## 3 Petal.Length 0.271 0.0278
## 4 Petal.Width 0.431 -0.110
#plot
ggplot()+
geom_point(data=sites_score,aes(x=CA1,y=CA2,color=iris[,5]))+
labs(x=paste("CA1 (",round(summary(model_ca)$cont$importance [2,1]*100,2)
,"% )",sep=""),
y=paste("CA2 (",round(summary(model_ca)$cont$importance [2,2]*100,2)
,"% )",sep=""))+
geom_segment(data=species_score,aes(x=0,y=0,xend=CA1,yend=CA2),size = 1,
arrow = arrow(length = unit(0.2, "inches")),color="steelblue")+
ggrepel::geom_text_repel(data=species_score,nudge_x = 0.4,
fontface="bold", color="black",
aes(x=CA1,y=CA2,label=rowname))+
stat_ellipse(type="norm",level= 0.9,
data=sites_score,aes(x=CA1,y=CA2,color=iris[,5]))+
theme_bw()
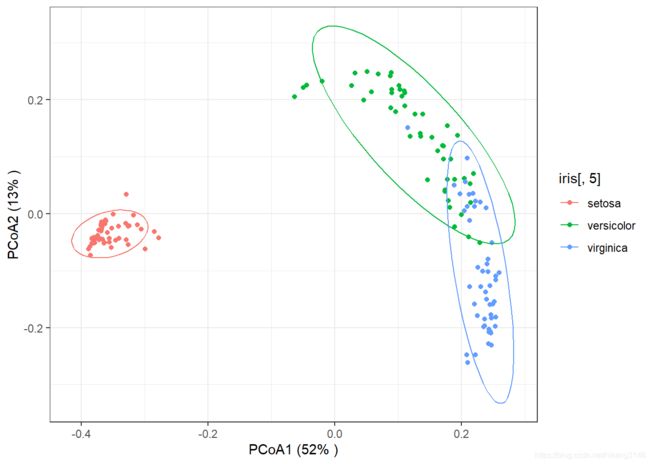
PCoA
PCoA可以采用其它距离矩阵(非欧式、卡方距离),如Bray-Cuurtis距离等。
#为防止出现大量负值特征值,需要对距离开方转换
model_pcoa<-vegdist(iris[,1:4],method="jaccard")%>% #计算距离
sqrt()%>% #平方根转换
cmdscale(k=2,eig=T) #多维标度转换
#变异解释
pcoa_explain<-round(model_pcoa$eig/sum(model_pcoa$eig)*100)
pcoa_explain
## [1] 52 13 5 4 3 2 2 2 1 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 0
## [28] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
## [55] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
## [82] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
## [109] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
## [136] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
#可视化
ggplot()+
geom_point(aes(x=model_pcoa$points[,1],
y=model_pcoa$points[,2],
color=iris[,5]))+
labs(x=paste("PCoA1 (",pcoa_explain[1],"% )",sep=""),
y=paste("PCoA2 (",pcoa_explain[2],"% )",sep=""))+
stat_ellipse(type="norm",level= 0.9,
aes(x=model_pcoa$points[,1],
y=model_pcoa$points[,2],color=iris[,5]))+
theme_bw()
NMDS
NMDS对距离采用rank排序的方式处理。
According to Clarke and Warwick 2001:
- stress < 0.05: excellent representation
- stress < 0.1: good representation
- stress < 0.2: acceptable representation
- stress > 0.3: unsatisfactory representation
model_nmds<-metaMDS(iris[,-5],distance="bray")
print(model_nmds$stress)
## [1] 0.03775523
# NMDS可视化
ggplot()+
geom_point(aes(x=model_nmds$points[,1],y=model_nmds$points[,2],color=iris[,5]))+
labs(x="NMDS1",y="NMDS2")+
annotate(geom='text',x=median(model_nmds$points[,2]),
y=max(model_nmds$points[,2]),
label=paste("stress=",round(model_nmds$stress,3),sep=""))+
stat_ellipse(type="norm",level= 0.9,
aes(x=model_nmds$points[,1],
y=model_nmds$points[,2],color=iris[,5]))+
theme_bw()
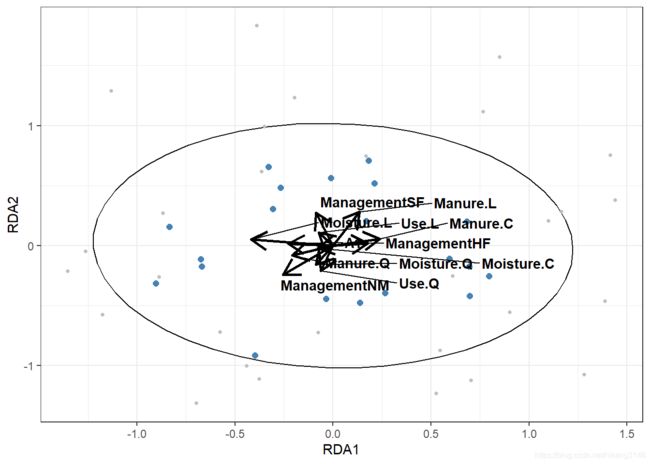
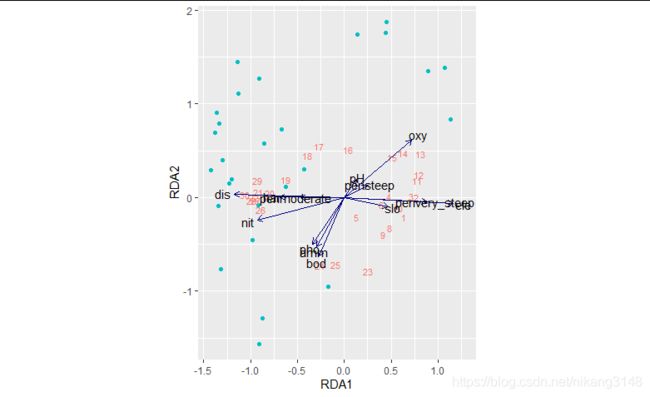
RDA约束性排序
RDA结合了线性回归与PCA主成分分析,是目前生态学常用的探索环境变量与群落组成关系的统计方法,具体分析原理可参考《Numerical Ecology》。对RDA的原理、操作已有很多文章介绍,此处不再赘言,本文着重在可视化部分
library(vegan)
data(dune)
data(dune.env)
model_rda<-rda(dune~.,data=dune.env,scale=T)
explain<-round(summary(eigenvals(model_rda))[2,1:2]*100) #explain%
rda_scores<-scores(model_rda,display=c("wa","sp","bp","cn"),scaling=1)
rda_scores$sites
rda_scores$species
RDA分析结果中最主要的是提取site、species、env的得分,可用scores()函数提取,分析结果也可用summary(rda_model)查看。
ggplot可视化
rda_scores_biplot<-rda_scores$biplot%>%as.data.frame()%>%rownames_to_column()%>%as_tibble()
ggplot()+
geom_point(data=as.data.frame(rda_scores$sites),
aes(x=RDA1,y=RDA2),color="steelblue",size=2)+
stat_ellipse(data=as.data.frame(rda_scores$sites),
aes(x=RDA1,y=RDA2),type="norm",level= 0.9)+
geom_point(data=as.data.frame(rda_scores$species),
aes(x=RDA1,y=RDA2),color="gray",size=1)+
geom_segment(data=as.data.frame(rda_scores_biplot),
aes(x=0,y=0,xend=RDA1,yend=RDA2),
size = 1,arrow = arrow(length = unit(0.2, "inches")),color="black")+
ggrepel::geom_text_repel(
data=rda_scores_biplot,
aes(x=RDA1,y=RDA2,label=rowname),
nudge_x = 0.4,fontface="bold", color="black")+
theme_bw()
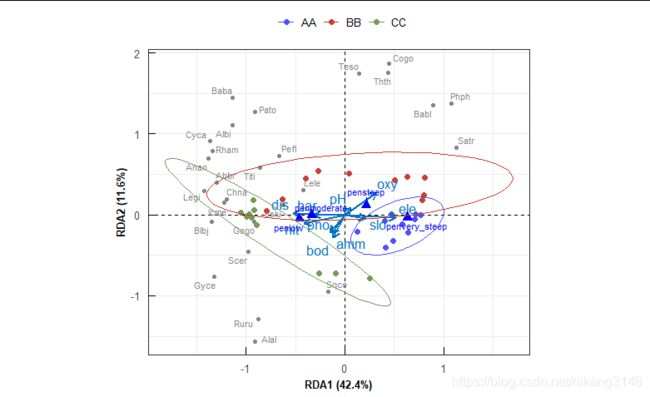
ggvegan可视化
比较方便的有ggvegan包(github安装),看着也还行,但是还是很多参数可调。
autoplot(spe.rda2,legend.position="none",
layers=c("sites","species","biplot"),
geom=c("text","point"),arrow=F,
scaling="sites"
)
rda.plot()函数
脱离ggvegan包,用ggplot绘图函数打包成函数,并用ggpubr包简化颜色设置。
rda.plot(spe.rda2,
scaling=1, #scaling code 1-3
display=c("sp","sites","env","cn"), #choose scores here
group=spe.tr, #treatments
palette = "igv", #see ggsci
envcol=get_palette("jco",8)[1], # envrionemtal variables see ggpubr
cncol="blue", #factors color
ellipse = T,
ellipse.type="norm",
ellipse.level=0.9
) #draw ellipe