K-means高维聚类与PCA降维
K-means高维与PCA降维
- K-means高维聚类
- PCA
查了很多博客,发现网上给出的K-means都是二维数据,没有高维示例,所以笔者将抄来的二维模板稍修改了一下,把Python实现的K-means高维聚类贴出来与大家共享,同时附一点PCA的实现(调用了三维作图,没有安包的可以删掉三维图)
K-means高维聚类
老师留的题目
Automatically determine the best cluster number K in k-means.
• Generate 1000 random N-Dimensional points;
• Try different K number;
• Compute SSE;
• Plot K-SSE figure;
• Choose the best K number (how to choose?).
Try different N number: 2, 3, 5, 10 Write the code in Jupyter Notebook Give me the screenshot of the code and results in Jupyter Notebook
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
N=2 #维度
def distance_fun(p1, p2, N):
result=0
for i in range(0,N):
result=result+((p1[i]-p2[i])**2)
return np.sqrt(result)
def mean_fun(a):
return np.mean(a,axis=0)
def farthest(center_arr, arr):
f = [0, 0]
max_d = 0
for e in arr:
d = 0
for i in range(center_arr.__len__()):
d = d + np.sqrt(distance_fun(center_arr[i], e, N))
if d > max_d:
max_d = d
f = e
return f
def closest(a, arr):
c = arr[1]
min_d = distance_fun(a, arr[1])
arr = arr[1:]
for e in arr:
d = distance_fun(a, e)
if d < min_d:
min_d = d
c = e
return c
if __name__=="__main__":
arr = np.random.randint(0,10000, size=(1000, 1, N))[:, 0, :] #1000个0-10000随机数
'''
block1= np.random.randint(0,2000, size=(100, 1, N))[:, 0, :] #分区间生成随机数
block2 = np.random.randint(2000,4000, size=(100, 1, N))[:, 0, :]
block3 = np.random.randint(4000,6000, size=(100, 1, N))[:, 0, :]
block4 = np.random.randint(6000,8000, size=(100, 1, N))[:, 0, :]
block5 = np.random.randint(8000,10000, size=(100, 1, N))[:, 0, :]
arr=np.vstack((block1,block2,block3,block4,block5))
'''
## 初始化聚类中心和聚类容器
K = 5
r = np.random.randint(arr.__len__() - 1)
center_arr = np.array([arr[r]])
cla_arr = [[]]
for i in range(K-1):
k = farthest(center_arr, arr)
center_arr = np.concatenate([center_arr, np.array([k])])
cla_arr.append([])
## 迭代聚类
n = 20
cla_temp = cla_arr
for i in range(n):
for e in arr:
ki = 0
min_d = distance_fun(e, center_arr[ki],N)
for j in range(1, center_arr.__len__()):
if distance_fun(e, center_arr[j],N) < min_d:
min_d = distance_fun(e, center_arr[j],N)
ki = j
cla_temp[ki].append(e)
for k in range(center_arr.__len__()):
if n - 1 == i:
break
center_arr[k] = mean_fun(cla_temp[k])
cla_temp[k] = []
if N>=2:
print(N,'维数据前两维投影')
col = ['gold', 'blue', 'violet', 'cyan', 'red','black','lime','brown','silver']
plt.figure(figsize=(10, 10))
for i in range(K):
plt.scatter(center_arr[i][0], center_arr[i][1], color=col[i])
plt.scatter([e[0] for e in cla_temp[i]], [e[1] for e in cla_temp[i]], color=col[i])
plt.show()
if N>=3:
print(N,'维数据前三维投影')
fig = plt.figure(figsize=(8, 8))
ax = Axes3D(fig)
for i in range(K):
ax.scatter(center_arr[i][0], center_arr[i][1], center_arr[i][2], color=col[i])
ax.scatter([e[0] for e in cla_temp[i]], [e[1] for e in cla_temp[i]],[e[2] for e in cla_temp[i]], color=col[i])
plt.show()
print(N,'维')
for i in range(K):
print('第',i+1,'个聚类中心坐标:')
for j in range(0,N):
print(center_arr[i][j])
首先解释一下代码,N为生成随机数字的维数,K为聚类中心数。
随机数我设定了两个部分,一个是0-10000完全随机数,另一个是0-10000均分成五个区间生成随机数。为什么要这么做呢?因为这样可以判别分类结果的正确与否。
如图
这是K=5时的完全随机数结果,可以看到初步分类正确

K=5时三维的完全随机数结果,可以看到初步分类正确
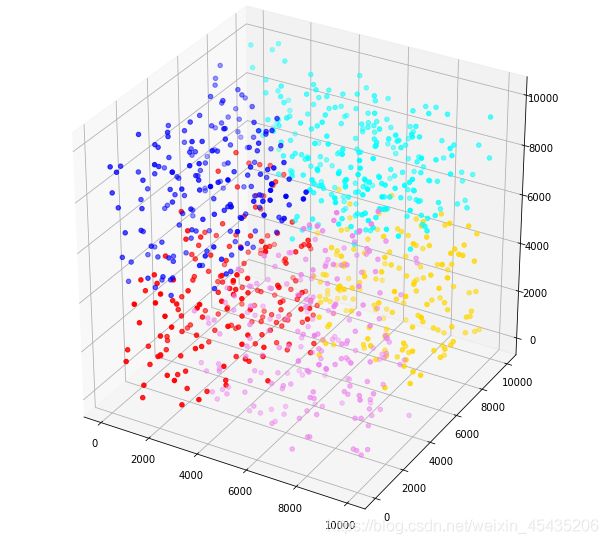
下面我们用区间生成随机数,这样可以直观地判别均值迭代好坏

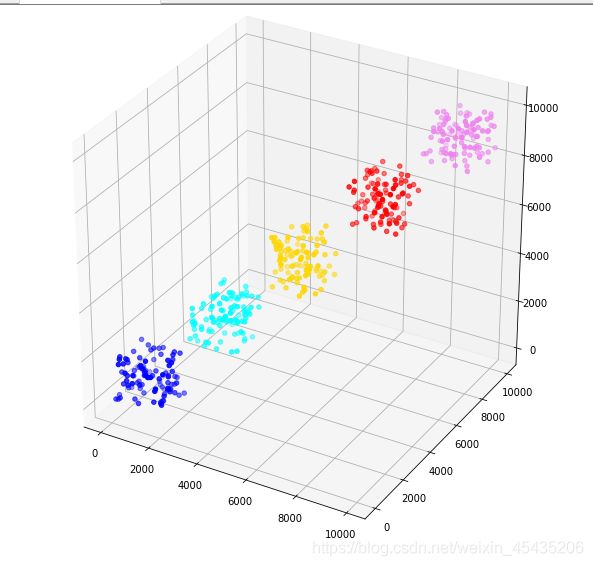
可以看到分区间产生的随机数能够非常好地展示出算法的准确性,说明迭代的效果是理想的。
下面我们来看更高维情况,如果N>=4时,图像没有办法直观显示了,我们只能依次取出N中的3维或2维做投影来观察。为了直接验证高维情况的正确与否,我们不再一张张地作图,而是直接判断聚类中心的坐标是否在随机数区间内。(此时就体现出分区间的随机数据及其重要)
更新补一下:SSE的实现
sse[i]=sse[i]+distance_fun(e, center_arr[ki],N)
在迭代中加个数组存放每次残差平方和数据
SSE图像大致如下:

很多时候会有小小的波折,暂时还没想清楚数学原理
PCA
题目
• Generate 5,00,000 random points with 200-D
• Dimension reduction to keep 90% energy using PCA
• Report how many dimensions are kept
• Compute k-means (k=100)
• Compare brute force NN and kd-tree, and report their running time
• Python, Jupyter Notebook
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
N=200 #维度
def distance_fun(p1, p2, N):
result=0
for i in range(0,N):
result=result+((p1[i]-p2[i])**2)
return np.sqrt(result)
def mean_fun(a):
return np.mean(a,axis=0)
def farthest(center_arr, arr):
f = [0, 0]
max_d = 0
for e in arr:
d = 0
for i in range(center_arr.__len__()):
d = d + np.sqrt(distance_fun(center_arr[i], e, N))
if d > max_d:
max_d = d
f = e
return f
def closest(a, arr):
c = arr[1]
min_d = distance_fun(a, arr[1])
arr = arr[1:]
for e in arr:
d = distance_fun(a, e)
if d < min_d:
min_d = d
c = e
return c
def pca(XMat):
average = mean_fun(XMat)
m, n = np.shape(XMat)
data_adjust = []
avgs = np.tile(average, (m, 1))
data_adjust = XMat - avgs
covX = np.cov(data_adjust.T) #计算协方差矩阵
featValue, featVec= np.linalg.eig(covX) #求解协方差矩阵的特征值和特征向量
index = np.argsort(-featValue) #依照featValue进行从大到小排序
sumfeatvalue=sum(index)
sumt=0
k=0
while(sumt<0.9*sumfeatvalue):
sumt+=index[k]
k+=1
finalData = []
selectVec = np.matrix(featVec.T[index]) #所以这里须要进行转置
finalData = data_adjust * selectVec.T
reconData = (finalData * selectVec) + average
return finalData, reconData,k
def plotBestFit(data1, data2):
dataArr1 = np.array(data1)
dataArr2 = np.array(data2)
m = np.shape(dataArr1)[0]
axis_x1 = []
axis_y1 = []
axis_x2 = []
axis_y2 = []
for i in range(m):
axis_x1.append(dataArr1[i,0])
axis_y1.append(dataArr1[i,1])
axis_x2.append(dataArr2[i,0])
axis_y2.append(dataArr2[i,1])
fig = plt.figure(figsize=(10, 10))
ax = fig.add_subplot(111)
#ax.scatter(axis_x1, axis_y1, s=50, c='red', marker='s')
ax.scatter(axis_x2, axis_y2, s=1, c='blue')
plt.show()
if __name__ == "__main__":
'''
arr = np.random.randint(0,10000, size=(1000, 1, N))[:, 0, :]
XMat=arr
'''
block1= np.random.randint(0,2000, size=(100000, 1, N))[:, 0, :] #分区间生成随机数
block2 = np.random.randint(2000,4000, size=(100000, 1, N))[:, 0, :]
block3 = np.random.randint(4000,6000, size=(100000, 1, N))[:, 0, :]
block4 = np.random.randint(6000,8000, size=(100000, 1, N))[:, 0, :]
block5 = np.random.randint(8000,10000, size=(100000, 1, N))[:, 0, :]
XMat=np.vstack((block1,block2,block3,block4,block5))
finalData, reconMat,pcaN = pca(XMat)
plotBestFit(finalData, reconMat) #输出前两维切片检查效果
print('降维到:',pcaN)
由于数据量很大,计算时间在几十秒左右,降维后200维大概到150维左右。PCA控制有两种方法,一种是根据特征值保留量计算应降维数,就像本题中的0.9。或者可以指定下降后维数,可以直接定量转换,但这种方法在实际中很不常用,因为当数据维度很高时,我们是无法直观的预知应降的维数的。代码中作图有两个部分,未偏移的降维数据和偏移后的降维数据,前者我注释掉了,可以跟据需求自己修改。
下面皮一下,200维降到二维看一下图像(只展示偏移后降维数据):

可以看到,分区间随机数的特征值基本还保留,效果蛮好,继续K-means也可以得到较好的结果,但这并不意味着我们可以直接指定下降后的维数。因为实际问题中的分类。远远比这简单的五个区间复杂,肆意指定维数会造成不可预计的数据丢失。
