多点取样的WES看肿瘤内部异质性文章合辑
肿瘤内部异质性问题已经是老生常谈了,在NGS如日中天的这些年,已经有非常多的多位点取样进行WES测序探索肿瘤异质性的(不同癌症研究的列表见文末),但是这个技术的可靠性并没有进行系统的评价,技术噪音,技术误差是可观存在的,所以就有了本研究:bioRxiv preprint first posted online Jan. 24, 2018; doi: http://dx.doi.org/10.1101/253195. 虽然并没有正式发布在SCI期刊,但是也值得解读。
3种不同的WES找somatic变异的策略
因为WES价格还是不低,所以很多研究并不好纳入肿瘤病人的配对正常组织进行测序,所以很有必要测试是否纳入肿瘤病人的配对正常组织进行测序的影响,所以作者测试了3个流程,包括:
- (i) the tumor DNA alone (i.e. tumor-only)
- (ii) tumor DNA and pooled unrelated normal DNA (i.e. cohort-normal)
- (iii) tumor DNA and patient matched normal DNA(i.e. matched-normal)
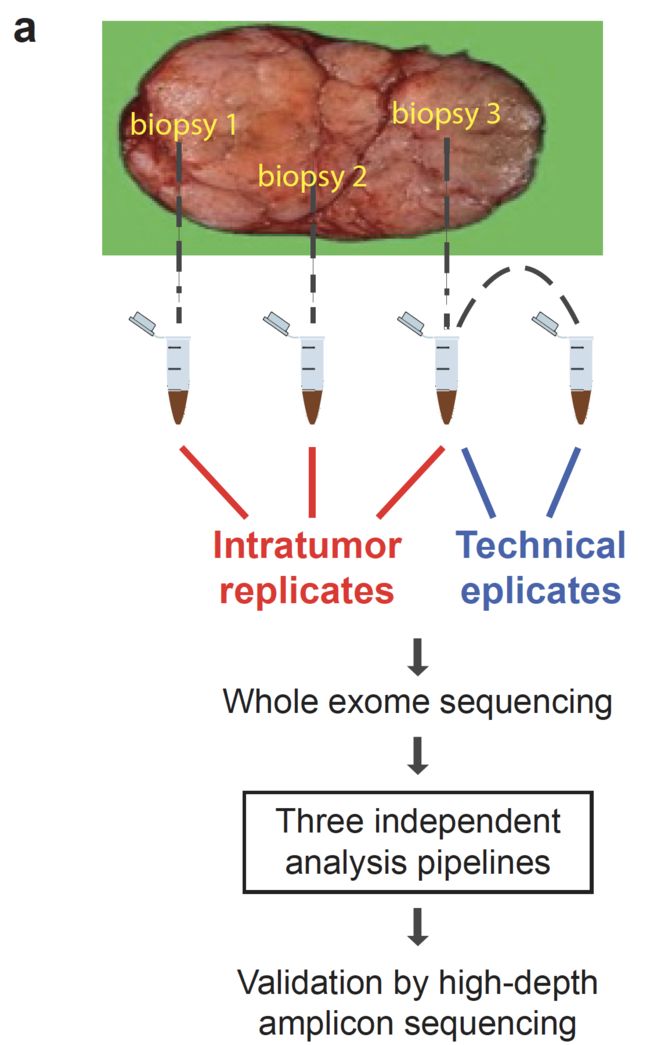
同时选择了6个乳腺癌病人,每个病人拿到3个肿瘤组织样品,同时也收集他们的normal组织进行测序,也就是比较标准的多位点取样进行WES测序探索肿瘤异质性的实验设计。流程图如下:
WES测序基本是: 160x (range 70x to 220x), 作者选择了所有流程找到的somatic变异位点合集再实验Iro torrent 平台的AmpliSeq方法进行平均605X的测序深度结果作为进标准。
根据金标准可以把突变进行分类,如下:
- true somatic
- absent
- germline-like (incorporating genuine germline variants and artifactual variant alleles caused by alignment biases and/or the sequencing technology)
- low-depth (i.e. technical failure with amplicon sequencing)
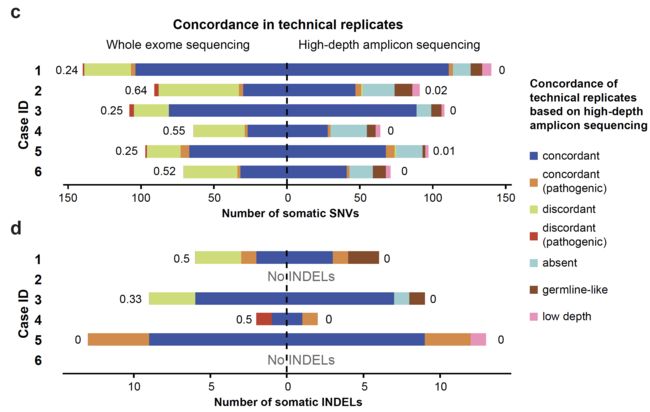
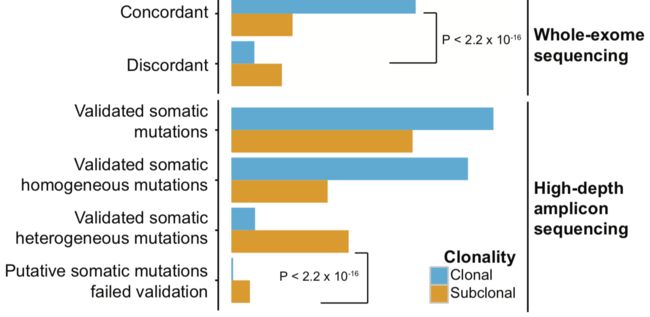
WES和高深度捕获测序的一致性
可以看到两个平台的一致性还不错。
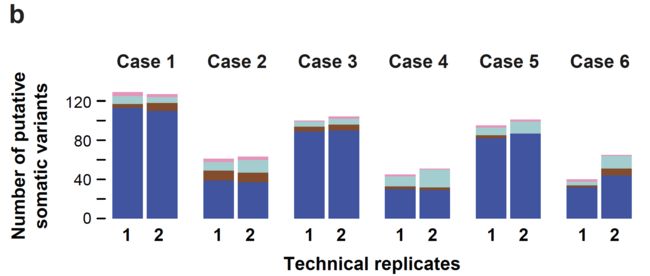
技术重复的一致性
可以看到同一个病人的2个技术重复的突变一致性很好,而且95% (range 73%-97%) 一致性的突变位点都是被证实的somatic位点,与之相比,那些不一致的位点,平均33% (range 14%-48%) 是somatic位点。
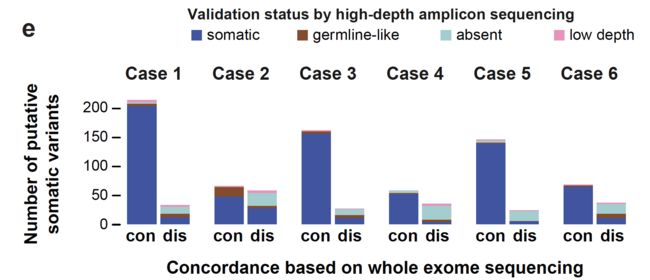
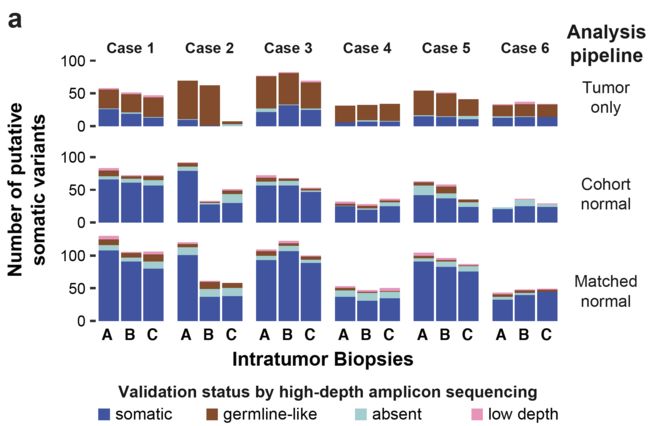
每个病人的somatic的SNV分类数量分布如下:
重要的结论是:一般测序深度下的WES其实是不足以完全检测到ITGH,尤其是在肿瘤纯度不够的时候。
3种数据处理流程的比较
首先从数量上看看这些流程的效果,开看到如果仅仅是使用tumor样品,得到的大多是germline位点,是很难找到真正的somatic突变的,如果使用一个normal数据集来进行过滤,效果会好一点,只有使用真正的配对的正常组织来进行过滤germline位点,最后得到的效果才最后!
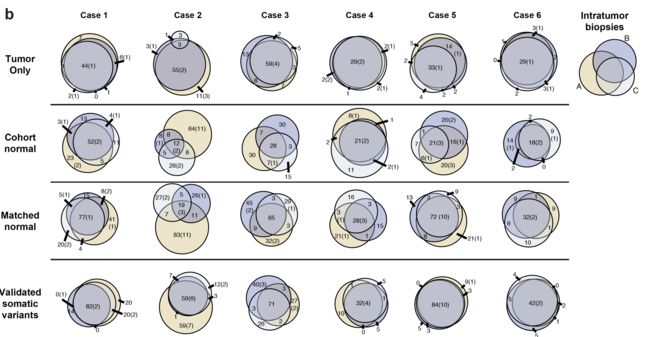
因为每个病人都是有3个部分的肿瘤样品进行测序,所以可以画韦恩图看看它们之间的overlap情况,如下:
可以看到大部分病人的不同部位的肿瘤样品之间的一致性非常好,但是有例外,作者并没有解释为什么。
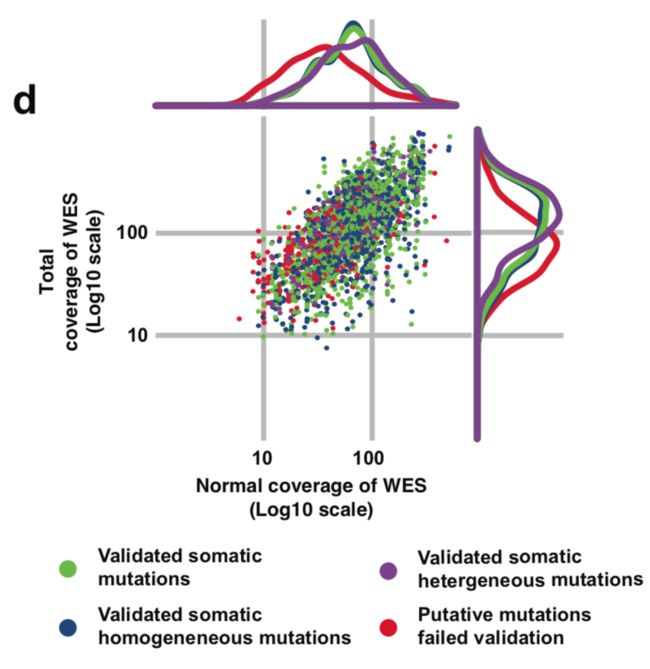
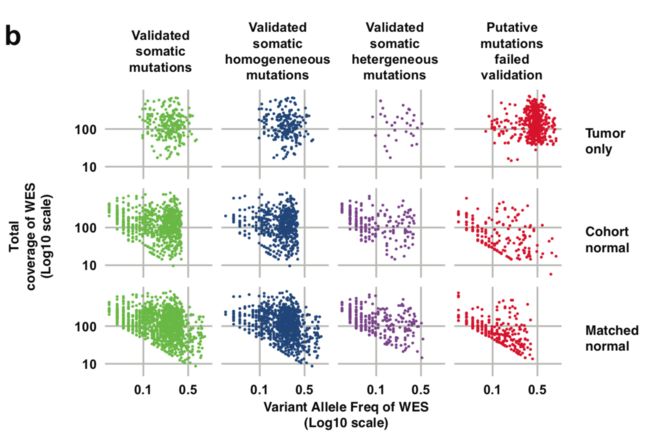
从测序深度来比较
这里作者选取了variant allele frequency (VAF) 和coverage来进行比较,图画的不错,首先看total coverage in the tumor (bottom) is plotted against the coverage in the matched normal sample of somatic mutations identified in all the specimens.
然后看 WES alternative allele coverage (all log10 scale) is plotted against VAF
区分真正的ITGH和WES技术误差
异质性很高的肿瘤含有很多亚克隆,它们决定着ITGH,所以作者使用 ABSOLUTE 来探索 WES的克隆,
需要考虑到4点才能识别真正的异质性突变位点,包括:
- VAF
- alternate allele depth
- total depth in tumor and normal
- mappability质量值
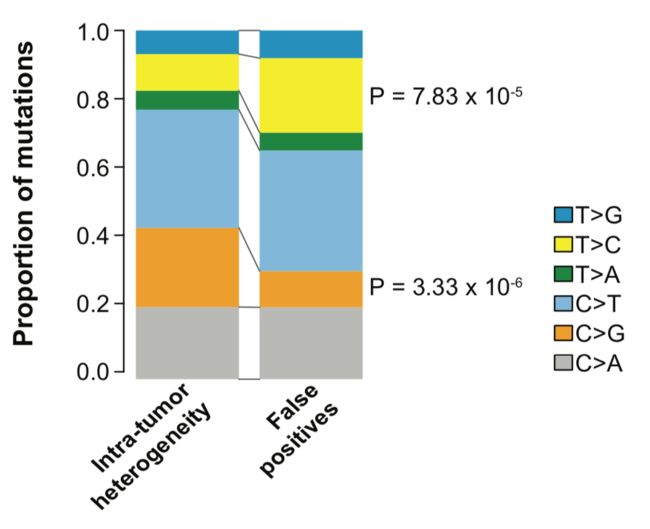
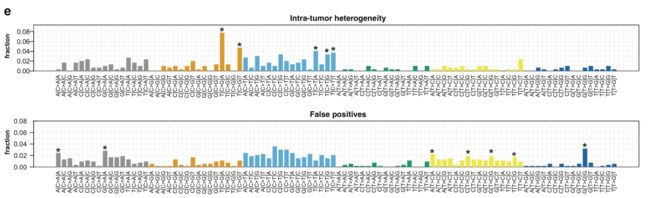
定义好真正造成肿瘤内部异质性的突变就可以跟那些假阳性的突变区分开来,然后就可以从6碱基突变形式和96碱基突变形式来比较这两群突变位点是否有特征区别,有非常显著的差异,如下:
同样,96碱基突变情况差异很显著:
讨论及展望
首先,本研究只纳入了6个病人的18个样品,而且iron torrent平台和illumina平台也有本质的区别,但仍然可以合理的得到结论,测序深度需要大于250X,可以一定程度的缓和假阳性和假阴性,尤其是在肿瘤纯度低于50%的情况下。总之,作者的研究,说明了平均测序深度184的时候肿瘤异质性被高估了,WES的技术误差被引入了ITGH。 然后就是去除那些位于比对质量值低的基因组区域的变异位点, 去除某些突变特征的是可以一定程度的降低假阳性假阴性,但是纯粹的执行测序深度的过滤是无效的。
最后是使用了多位点区域WES技术的文章列表:
1.Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med.366, 883–892 (2012).
PubMed Central
2.Gerlinger, M. et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 46, 225–233 (2014).
PubMed Central
3.Yates, L. R. et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat. Med. 21, 751–759 (2015). 样本量很可观:50 patients’ tumors (total 303) 有WGS和捕获panel测序两种形式。
PubMed Central
4.Stachler, M. D. et al. Paired exome analysis of Barrett’s esophagus and adenocarcinoma. Nat. Genet. 47, 1047–1055 (2015).
PubMed Central
5.Murugaesu, N. et al. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov. 5, 821–831 (2015).
PubMed Central
6.Zhang, J. et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science346, 256–259 (2014).
PubMed Central
7.de Bruin, E. C. et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346, 251–256 (2014).
PubMed Central
8.Schwarz, R. F. et al. Spatial and temporal heterogeneity in high-grade serous ovarian cancer: a phylogenetic analysis. PLoS Med.12, e1001789 (2015).
PubMed Central
9.Haffner, M. C. et al. Tracking the clonal origin of lethal prostate cancer. J. Clin. Invest. 123, 4918–4922 (2013).
PubMed Central
10.Gundem, G. et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015).PubMed Central
11.Makohon-Moore, A. P. et al. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat. Genet. 49, 358–366 (2017). PubMed Central
12.Volume 19, May 2017, Intratumor Heterogeneity in Primary Kidney Cancer Revealed by Metabolic Profiling of Multiple Spatially Separated Samples within Tumors
还有:Published: 23 July 2018 A temporal shift of the evolutionary principle shaping intratumor heterogeneity in colorectal cancer 日本研究团队成果,
还有:April 2017 Multi-Region Exome Sequencing Reveals the Clonal Evolution of Colitis-Associated Colorectal Cancer
首先是文章列表:
1.Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med.366, 883–892 (2012).
PubMed Central
2.Gerlinger, M. et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 46, 225–233 (2014).
PubMed Central
3.Yates, L. R. et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat. Med. 21, 751–759 (2015). 样本量很可观:50 patients’ tumors (total 303) 有WGS和捕获panel测序两种形式。
PubMed Central
4.Stachler, M. D. et al. Paired exome analysis of Barrett’s esophagus and adenocarcinoma. Nat. Genet. 47, 1047–1055 (2015).
PubMed Central
5.Murugaesu, N. et al. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov. 5, 821–831 (2015).
PubMed Central
6.Zhang, J. et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science346, 256–259 (2014).
PubMed Central
7.de Bruin, E. C. et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346, 251–256 (2014).
PubMed Central
8.Schwarz, R. F. et al. Spatial and temporal heterogeneity in high-grade serous ovarian cancer: a phylogenetic analysis. PLoS Med.12, e1001789 (2015).
PubMed Central
9.Haffner, M. C. et al. Tracking the clonal origin of lethal prostate cancer. J. Clin. Invest. 123, 4918–4922 (2013).
PubMed Central
10.Gundem, G. et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015).PubMed Central
11.Makohon-Moore, A. P. et al. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat. Genet. 49, 358–366 (2017). PubMed Central
- Volume 19, May 2017, Intratumor Heterogeneity in Primary Kidney Cancer Revealed by Metabolic Profiling of Multiple Spatially Separated Samples within Tumors
还有:Published: 23 July 2018 A temporal shift of the evolutionary principle shaping intratumor heterogeneity in colorectal cancer 日本研究团队成果,
还有:April 2017 Multi-Region Exome Sequencing Reveals the Clonal Evolution of Colitis-Associated Colorectal Cancer
(文章转自jimmy的2018年阅读文献笔记)
生信基础知识大全系列:生信基础知识100讲
史上最强的生信自学环境准备课来啦!! 7次改版,11节课程,14K的讲稿,30个夜晚打磨,100页PPT的课程。
如果需要组装自己的服务器;代办生物信息学服务器
如果需要帮忙下载海外数据(GEO/TCGA/GTEx等等),点我?
如果需要线下辅导及培训,看招学徒
如果需要个人电脑:个人计算机推荐
如果需要置办生物信息学书籍,看:生信人必备书单
如果需要实习岗位:实习职位发布
如果需要售后:点我