xgboost2 以及使用XGB.CV来进行调参
#xgboost的几种弱评估器,booster
for booster in ["gbtree","gblinear","dart"]:
reg=XGBR(n_estimatiors=22,learning_rate=0.1
,rangdom_state=420,booster=booster).fit(Xtrain,ytrain)
print(booster)
print(reg.score(Xtest,ytest))
#查看XGBOOST本身的调用方式,与sklearn方法不同
#首先是使用sklearn库调用的方式
from xgboost import XGBRegressor as XGBR
reg=XGBR(n_estimators=22,random_state=420).fit(Xtrain,ytrain)
reg.score(Xtest,ytest)
MSE(ytest,reg.predict(Xtest))
#接下来使用xgboost本身的库来调用
import xgboost as xgb
#使用DMatrix读取数据
dtrain=xgb.DMatrix(Xtrain,ytrain)#注意需要把标签一同穿进去
dtest=xgb.DMatrix(Xtest,ytest)
param={"silent":False,
"objective":"reg:linear",
"eta":0.1}
num_round=22#这个参数放外面,是因为可以读的
#param里的不能写外面
bst=xgb.train(param,dtrain,num_round)
preds=bst.predict(dtest)
from sklearn.metrics import r2_score
r2_score(ytest,preds)
MSE(ytest,preds)
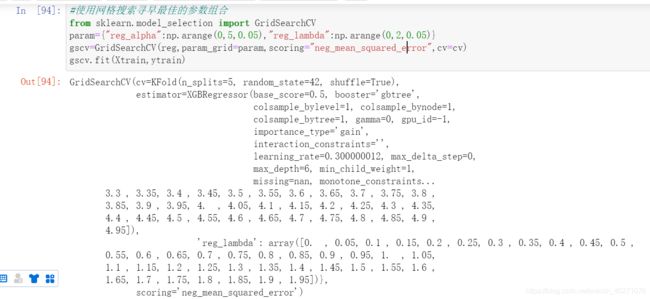
#使用网格搜索寻早最佳的参数组合
from sklearn.model_selection import GridSearchCV
param={"reg_alpha":np.arange(0,5,0.05),"reg_lambda":np.arange(0,2,0.05)}
gscv=GridSearchCV(reg,param_grid=param,scoring="neg_mean_squared_error",cv=cv)
gscv.fit(Xtrain,ytrain)
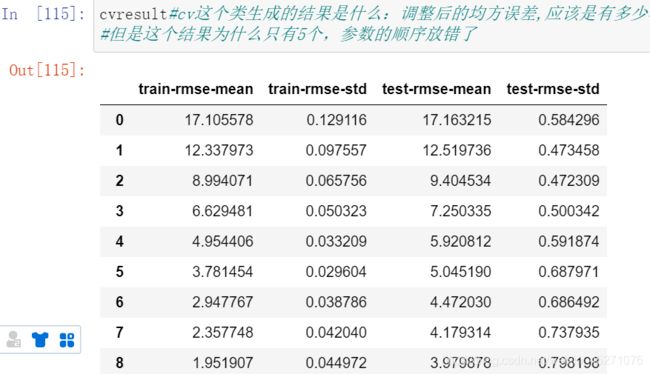
#使用xgb.cv这个类来调整r这个参数,gamma是用来控制让树停止生长得
import xgboost as xgb
dfull=xgb.DMatrix(X,y)#使用全数据
param={"silent":True,"obj":"reg:linear","gamma":0}
num_round=22
n_fold=5
time0=time()
cvresult=xgb.cv(param,dfull,num_round,n_fold)#这个里面参数的顺序不能错
print(datetime.datetime.fromtimestamp(time()-time0).strftime("%M%S%f"))
#使用xgb.cv这个类来调整r这个参数,gamma是用来控制让树停止生长得
#修改模型评估指标
import xgboost as xgb
dfull=xgb.DMatrix(X,y)#使用全数据
param={"silent":True,"obj":"reg:linear","gamma":0,"eval_metricc":"mae"}#修改模型评估指标
num_round=22
n_fold=5
time0=time()
cvresult=xgb.cv(param,dfull,num_round,n_fold)#这个里面参数的顺序不能错
#print(datetime.datetime.fromtimestamp(time()-time0).strftime("%M%S%f"))
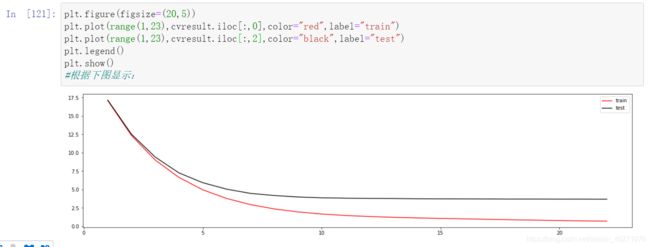
plt.figure(figsize=(20,5))
plt.plot(range(1,23),cvresult.iloc[:,0],color="red",label="train")
plt.plot(range(1,23),cvresult.iloc[:,2],color="black",label="test")
plt.legend()
plt.show()
#根据上图可以看到:最佳的n_estimators,因为后面都是平缓的了
#现在模型处于过拟合,一种是把训练集红色线往上,一种是让测试集黑线下降
#那么从现有图来看,两条线越接近,就是我们调参的目标
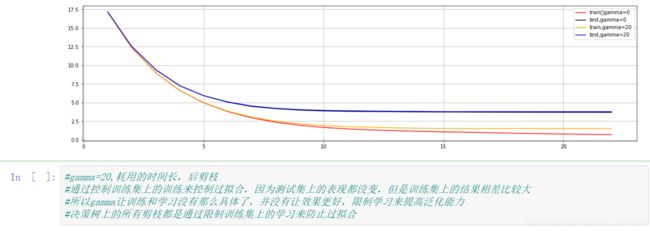
#接下来如何调参,修改gamma值,gamma越大,算法就越保守,树的叶子数量就越少,模型的复杂的就越低,就越不可能过拟合
import xgboost as xgb
dfull=xgb.DMatrix(X,y)#使用全数据
param1={"silent":True,"obj":"reg:linear","gamma":0,"eval_metricc":"mae"}#修改模型评估指标
param2={"silent":True,"obj":"reg:linear","gamma":20,"eval_metricc":"mae"}#修改模型评估指标
num_round=22
n_fold=5
time0=time()
cvresult1=xgb.cv(param1,dfull,num_round,n_fold)#这个里面参数的顺序不能错
#print(datetime.datetime.fromtimestamp(time()-time0).strftime("%M%S%f"))
cvresult2=xgb.cv(param2,dfull,num_round,n_fold)
plt.figure(figsize=(20,5))
plt.grid()
plt.plot(range(1,23),cvresult1.iloc[:,0],color="red",label="train,gamma=0")
plt.plot(range(1,23),cvresult1.iloc[:,2],color="black",label="test,gamma=0")
plt.plot(range(1,23),cvresult2.iloc[:,0],color="orange",label="train,gamma=20")
plt.plot(range(1,23),cvresult2.iloc[:,2],color="blue",label="test,gamma=20")
plt.legend()
plt.show()
#分类举例,使用乳腺癌数据
from sklearn.datasets import load_breast_cancer
data=load_breast_cancer()
X=data.data
y=data.target
dfull=xgb.DMatrix(X,y)
param1={"silent":True,"obj":"binary:logistic","gamma":0,"n_fold":5}
param2={"silent":True,"obj":"binary:logistic","gamma":2,"n_fold":5}
num_round=100
cvresult1=xgb.cv(param1,dfull,num_round)
cvresult2=xgb.cv(param2,dfull,num_round)
plt.figure(figsize=(20,5))
plt.grid()
plt.plot(range(1,101),cvresult1.iloc[:,0],color="red",label="train,gamma=0")
plt.plot(range(1,101),cvresult1.iloc[:,2],color="black",label="test,gamma=0")
plt.plot(range(1,101),cvresult2.iloc[:,0],color="blue",label="train,gamma=2")
plt.plot(range(1,101),cvresult2.iloc[:,2],color="green",label="test,gamma=2")
plt.legend()
plt.show()
#也就是说虽然测试集和训练集的距离确实小了,但是测试集的效果也差了
#训练集和测试集的效果都降低了
#可能的原因是gamma调整的太大了,那么改小gamma看一下结果
from sklearn.datasets import load_breast_cancer
data=load_breast_cancer()
X=data.data
y=data.target
dfull=xgb.DMatrix(X,y)
param1={"silent":True,"obj":"binary:logistic","gamma":0,"n_fold":5}
param2={"silent":True,"obj":"binary:logistic","gamma":1,"n_fold":5}
num_round=100
cvresult1=xgb.cv(param1,dfull,num_round)
cvresult2=xgb.cv(param2,dfull,num_round)
plt.figure(figsize=(20,5))
plt.grid()
plt.plot(range(1,101),cvresult1.iloc[:,0],color="red",label="train,gamma=0")
plt.plot(range(1,101),cvresult1.iloc[:,2],color="black",label="test,gamma=0")
plt.plot(range(1,101),cvresult2.iloc[:,0],color="blue",label="train,gamma=1")
plt.plot(range(1,101),cvresult2.iloc[:,2],color="green",label="test,gamma=1")
plt.legend()
plt.show()