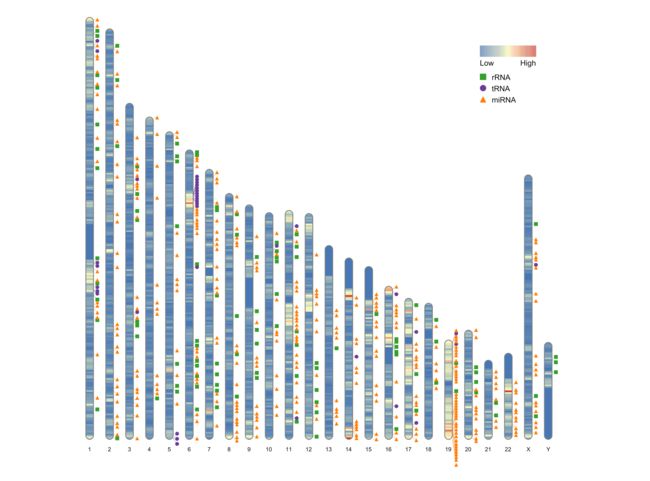
已有的工具如circos, 只能绘制弯的染色体,又如R包chromoMap,IdeoViz,karyploteR,ggbio和在线工具Idiographica,虽然能绘制直的染色体,但仅支持人,小鼠,大鼠和果蝇等几个物种,不支持自定义物种,不够自由。R包RIdeogram可以可视化染色体上的全基因组数据,并且输出SVG格式的文件,也可以将SVG格式的文件转化为pdf, png, tiff, 或jpg格式。
安装
install.packages('RIdeogram')
require(RIdeogram)
输入文件
data(human_karyotype, package="RIdeogram")
data(gene_density, package="RIdeogram")
data(Random_RNAs_500, package="RIdeogram")
head(human_karyotype)
#> Chr Start End CE_start CE_end
#> 1 1 0 248956422 122026459 124932724
#> 2 2 0 242193529 92188145 94090557
#> 3 3 0 198295559 90772458 93655574
#> 4 4 0 190214555 49712061 51743951
#> 5 5 0 181538259 46485900 50059807
#> 6 6 0 170805979 58553888 59829934
karyotype文件:可以是五列(包含中心粒位置)或三列(不含中心粒位置)
第一列:染色体号
第二列:起始
第三列:终止
第四列:中心粒起始位置
第五列:中心粒终止位置
head(gene_density)
#> Chr Start End Value
#> 1 1 1 1000000 65
#> 2 1 1000001 2000000 76
#> 3 1 2000001 3000000 35
#> 4 1 3000001 4000000 30
#> 5 1 4000001 5000000 10
#> 6 1 5000001 6000000 10
基因密度文件:
第一列:染色体号
第二列:起始
第三列:终止
第四列:基因密度值
head(Random_RNAs_500)
#> Type Shape Chr Start End color
#> 1 tRNA circle 6 69204486 69204568 6a3d9a
#> 2 rRNA box 3 68882967 68883091 33a02c
#> 3 rRNA box 5 55777469 55777587 33a02c
#> 4 rRNA box 21 25202207 25202315 33a02c
#> 5 miRNA triangle 1 86357632 86357687 ff7f00
#> 6 miRNA triangle 11 74399237 74399333 ff7f00
染色体旁的标记文件:
第一列:标记类型
第二列:标记形状
第三列:染色体号
第四列:起始
第五列:终止
第六列:颜色
也可以加载自己的数据。
karyotype <- read.table("karyotype.txt", sep = "\t", header = T, stringsAsFactors = F)
density <- read.table("data_1.txt", sep = "\t", header = T, stringsAsFactors = F)
label <- read.table("data_2.txt", sep = "\t", header = T, stringsAsFactors = F)
另外,该R包中还提供了一个GFFex函数用来从GFF文件中提取绘制染色体上热图的信息(例如基因密度)。
首先,需要准备物种的karyotype文件,格式与上述的相同,且保证第一列染色体号与GFF文件中的相同。然后,使用GFFex提取基因密度信息。
gene_density <- GFFex(input = "gencode.v32.annotation.gff3.gz", karyotype = "human_karyotype.txt", feature = "gene", window = 1000000)
其中,feature选项可以改为要绘制的特征类型,window选项可以更改统计的窗口大小。
用法
基本染色体绘制
ideogram(karyotype = human_karyotype)
convertSVG("chromosome.svg", device = "png")
基因密度热图绘制
ideogram(karyotype = human_karyotype, overlaid = gene_density)
convertSVG("chromosome.svg", device = "png")
标记类型绘制
ideogram(karyotype = human_karyotype, label = Random_RNAs_500, label_type = "marker")
convertSVG("chromosome.svg", device = "png")
染色体,基因密度和标记同时绘制
ideogram(karyotype = human_karyotype, overlaid = gene_density, label = Random_RNAs_500, label_type = "marker")
convertSVG("chromosome.svg", device = "png")
修改基因密度热图的颜色
ideogram(karyotype = human_karyotype, overlaid = gene_density, label = Random_RNAs_500, label_type = "marker", colorset1 = c("#fc8d59", "#ffffbf", "#91bfdb"))
convertSVG("chromosome.svg", device = "png")
无中心粒染色体的绘制
human_karyotype <- human_karyotype[,1:3]
ideogram(karyotype = human_karyotype, overlaid = gene_density, label = Random_RNAs_500, label_type = "marker")
convertSVG("chromosome.svg", device = "png")
修改染色体宽度值(染色体数较少时)
# default width is 170
human_karyotype <- human_karyotype[1:10,]
ideogram(karyotype = human_karyotype, overlaid = gene_density, label = Random_RNAs_500, label_type = "marker")
convertSVG("chromosome.svg", device = "png")
# change width to 100
human_karyotype <- human_karyotype[1:10,]
ideogram(karyotype = human_karyotype, overlaid = gene_density, label = Random_RNAs_500, label_type = "marker", width = 100)
convertSVG("chromosome.svg", device = "png")
移动图例位置
# change Lx and Ly to 80, 25
ideogram(karyotype = human_karyotype, overlaid = gene_density, label = Random_RNAs_500, label_type = "marker", width = 100, Lx = 80, Ly = 25)
convertSVG("chromosome.svg", device = "png")
绘制热图标签
data(human_karyotype, package="RIdeogram") #reload the karyotype data
ideogram(karyotype = human_karyotype, overlaid = gene_density, label = LTR_density, label_type = "heatmap", colorset1 = c("#f7f7f7", "#e34a33"), colorset2 = c("#f7f7f7", "#2c7fb8")) #use the arguments 'colorset1' and 'colorset2' to set the colors for gene and LTR heatmaps, separately.
convertSVG("chromosome.svg", device = "png")
绘制线形标签
# 单线形标签
data(liriodendron_karyotype, package="RIdeogram") #load the karyotype data
data(Fst_between_CE_and_CW, package="RIdeogram") #load the Fst data for overlaid heatmap
data(Pi_for_CE, package="RIdeogram") #load the Pi data for one-line label
head(Pi_for_CE) #this data has a similar format with the heatmap data with additional column of "Color" which indicate the color for the line.
#> Chr Start End Value Color
#> 1 1 1 2000000 0.00273566 fc8d62
#> 2 1 1000001 3000000 0.00239580 fc8d62
#> 3 1 2000001 4000000 0.00319407 fc8d62
#> 4 1 3000001 5000000 0.00286900 fc8d62
#> 5 1 4000001 6000000 0.00186596 fc8d62
#> 6 1 5000001 7000000 0.00186182 fc8d62
ideogram(karyotype = liriodendron_karyotype, overlaid = Fst_between_CE_and_CW, label = Pi_for_CE, label_type = "line", colorset1 = c("#e5f5f9", "#99d8c9", "#2ca25f"))
convertSVG("chromosome.svg", device = "png")
# 双线形标签
data(liriodendron_karyotype, package="RIdeogram") #load the karyotype data
data(Fst_between_CE_and_CW, package="RIdeogram") #load the Fst data for overlaid heatmap
data(Pi_for_CE_and_CW, package="RIdeogram") #load the Pi data for two-line label
head(Pi_for_CE_and_CW) #this data has a similar format with the one for one-line label with additional two columns indicating the second feature you want to show. When you prepare your own data, please keep the exact same column names.
#> Chr Start End Value_1 Color_1 Value_2 Color_2
#> 1 1 1 2000000 0.00273566 fc8d62 0.00385702 8da0cb
#> 2 1 1000001 3000000 0.00239580 fc8d62 0.00331109 8da0cb
#> 3 1 2000001 4000000 0.00319407 fc8d62 0.00374530 8da0cb
#> 4 1 3000001 5000000 0.00286900 fc8d62 0.00339141 8da0cb
#> 5 1 4000001 6000000 0.00186596 fc8d62 0.00305246 8da0cb
#> 6 1 5000001 7000000 0.00186182 fc8d62 0.00323655 8da0cb
ideogram(karyotype = liriodendron_karyotype, overlaid = Fst_between_CE_and_CW, label = Pi_for_CE_and_CW, label_type = "line", colorset1 = c("#e5f5f9", "#99d8c9", "#2ca25f"))
convertSVG("chromosome.svg", device = "png")
绘制多边形标签
# 单多边形标签
data(liriodendron_karyotype, package="RIdeogram") #load the karyotype data
data(Fst_between_CE_and_CW, package="RIdeogram") #load the Fst data for overlaid heatmap
data(Pi_for_CE, package="RIdeogram") #load the Pi data for one-polygon label
ideogram(karyotype = liriodendron_karyotype, overlaid = Fst_between_CE_and_CW, label = Pi_for_CE, label_type = "polygon", colorset1 = c("#e5f5f9", "#99d8c9", "#2ca25f"))
convertSVG("chromosome.svg", device = "png")
# 双多边形标签
data(liriodendron_karyotype, package="RIdeogram") #load the karyotype data
data(Fst_between_CE_and_CW, package="RIdeogram") #load the Fst data for overlaid heatmap
data(Pi_for_CE_and_CW, package="RIdeogram") #load the Pi data for two-polygon label
ideogram(karyotype = liriodendron_karyotype, overlaid = Fst_between_CE_and_CW, label = Pi_for_CE_and_CW, label_type = "polygon", colorset1 = c("#e5f5f9", "#99d8c9", "#2ca25f"))
convertSVG("chromosome.svg", device = "png")
此外,还可以修改device参数来转换图片为其他格式,比如tiff, pdf, jpg等,还可以修改dpi参数来设置图片的分辨率(默认为300).
convertSVG("chromosome.svg", device = "tiff", dpi = 600)
该R包中还提供了四个快捷方式进行图片格式转换。
svg2tiff("chromosome.svg")
svg2pdf("chromosome.svg")
svg2jpg("chromosome.svg")
svg2png("chromosome.svg")
基因组共线性区域可视化
两个基因组间共线性区域的绘制,
data(karyotype_dual_comparison, package="RIdeogram")
head(karyotype_dual_comparison)
#> Chr Start End fill species size color
#> 1 I 1 23037639 969696 Grape 12 252525
#> 2 II 1 18779884 969696 Grape 12 252525
#> 3 III 1 19341862 969696 Grape 12 252525
#> 4 IV 1 23867706 969696 Grape 12 252525
#> 5 V 1 25021643 969696 Grape 12 252525
#> 6 VI 1 21508407 0ab276 Grape 12 252525
table(karyotype_dual_comparison$species)
#>
#> Grape Populus
#> 19 19
data(synteny_dual_comparison, package="RIdeogram")
head(synteny_dual_comparison)
#> Species_1 Start_1 End_1 Species_2 Start_2 End_2 fill
#> 1 1 12226377 12267836 2 5900307 5827251 cccccc
#> 2 15 5635667 5667377 17 4459512 4393226 cccccc
#> 3 9 7916366 7945659 3 8618518 8486865 cccccc
#> 4 2 8214553 8242202 18 5964233 6027199 cccccc
#> 5 13 2330522 2356593 14 6224069 6138821 cccccc
#> 6 11 10861038 10886821 10 8099058 8011502 cccccc
karyotype_dual_comparison文件格式
Chr: 染色体号
Start: 起始
End: 终止
fill: 染色体填充色
species:物种名
size: 物种名字体大小
color: 物种名字体颜色
synteny_dual_comparison文件格式
Species_1:物种1染色体号
Start_1,End_1:物种1染色体区域位置
Species_2:物种2染色体号
Start_2,End_2:物种2染色体区域位置
ideogram(karyotype = karyotype_dual_comparison, synteny = synteny_dual_comparison)
convertSVG("chromosome.svg", device = "png")
三个基因组间共线性区域的绘制,
data(karyotype_ternary_comparison, package="RIdeogram")
head(karyotype_ternary_comparison)
#> Chr Start End fill species size color
#> 1 NA 1 15980527 fcb06b Amborella 10 fcb06b
#> 2 NA 1 11522362 fcb06b Amborella 10 fcb06b
#> 3 NA 1 11085951 fcb06b Amborella 10 fcb06b
#> 4 NA 1 10537363 fcb06b Amborella 10 fcb06b
#> 5 NA 1 9585472 fcb06b Amborella 10 fcb06b
#> 6 NA 1 9414115 fcb06b Amborella 10 fcb06b
table(karyotype_ternary_comparison$species)
#>
#> Amborella Grape Liriodendron
#> 100 19 19
data(synteny_ternary_comparison, package="RIdeogram")
head(synteny_ternary_comparison)
#> Species_1 Start_2 End_2 Species_2 Start_1 End_1 fill type
#> 1 1 4761181 2609697 1 342802 981451 cccccc 1
#> 2 6 6344197 8074393 1 15387184 16716190 cccccc 1
#> 3 10 6457890 9052487 1 11224953 14959548 cccccc 1
#> 4 13 6318795 1295413 1 20564870 21386271 cccccc 1
#> 5 16 1398101 2884119 1 21108654 22221088 cccccc 1
#> 6 16 1482529 2093625 1 21864494 22364888 cccccc 1
tail(synteny_ternary_comparison, n = 20)
#> Species_1 Start_2 End_2 Species_2 Start_1 End_1 fill type
#> 571 16 19278042 20828694 2 95267449 93334736 cccccc 3
#> 572 12 20546006 22461088 2 22647943 18365764 cccccc 3
#> 573 4 22259262 23453956 2 15068249 17839485 cccccc 3
#> 574 14 22377895 23821929 2 97299880 96033346 cccccc 3
#> 575 6 1538773 2808373 1 91285578 95681546 cccccc 3
#> 576 11 3381792 4954528 1 67689752 75286468 cccccc 3
#> 577 9 4814481 6975840 1 69506847 76015710 cccccc 3
#> 578 10 7091825 9742616 1 19333526 24516133 cccccc 3
#> 579 13 22063957 23402389 1 95843870 92195256 cccccc 3
#> 580 7 679765 1881756 6 7365421 7531534 e41a1c 1
#> 581 7 679765 2752867 13 501561 766473 e41a1c 1
#> 582 7 679765 3012501 8 7406703 8222490 e41a1c 1
#> 583 7 2049369 2942034 14 29350547 34369929 e41a1c 2
#> 584 7 2075095 1538540 10 28985737 30815217 e41a1c 2
#> 585 13 531939 834472 14 28866243 35278211 e41a1c 3
#> 586 8 7427221 8894821 14 28632063 34805893 e41a1c 3
#> 587 6 7567597 7690342 14 32050301 34913801 e41a1c 3
#> 588 13 501561 876423 10 30496700 27874100 e41a1c 3
#> 589 6 7171014 7815454 10 31408837 27660041 e41a1c 3
#> 590 8 5773528 9346871 10 31408837 26585934 e41a1c 3
ideogram(karyotype = karyotype_ternary_comparison, synteny = synteny_ternary_comparison)
convertSVG("chromosome.svg", device = "png")
更好的阅读体验请移步这里>>
参考
- RIdeogram: drawing SVG graphics to visualize and map genome-wide data on idiograms
- RIdeogram Github