主要学习内容:Linux系统的软件安装
第一步:登陆服务器(不用再登阿里云啦,还是不大理解我电脑里的的putty相当于什么,可以理解为是用来登陆服务器的助手吗,服务器是大家可以一起用但又互不干扰的一个超级大的计算机,然后这个计算机安的系统就是linux的。)
第二步:bzip2(理解为一款解压软件,给你的linux系统的电脑先安上它,再下载别的东西安装。)
bzip2
-bash: bzip2: command not found
yum install -y bzip2
Installed:
bzip2.x86_64 0:1.0.6-13.el7
Complete!
第三步:安装和配置miniconda (再给你电脑安个应用商店,(miniconda——conda的精华版miniconda)
官网下载python3.6(计算机程序设计语言)对应的版本
复制网址(sh可能就当作exe)
下载
wget https://repo.continuum.io/miniconda/Miniconda3-latest-Linux-x86_64.sh
2018-08-22 21:18:21 (7.00 MB/s) - ‘Miniconda3-latest-Linux-x86_64.sh’ saved [58468498/58468498]
安装
bash Miniconda3-latest-Linux-x86_64.sh
enter
Do you accept the license terms? [yes|no]
[no] >>>
Please answer 'yes' or 'no':'
>>> yes
enter
installation finished.
Do you wish the installer to prepend the Miniconda3 install location
to PATH in your /root/.bashrc ? [yes|no]
[no] >>> yes
Thank you for installing Miniconda3!
激活
source ~/.bashrc
[root@shanshuang121 biosofe]# conda
命令行([root@shanshuang121 biosofe]# 号后面就是)
添加国内镜像
conda config --add channels https://mirrors.ustc.edu.cn/anaconda/pkgs/free
conda config --add channels https://mirrors.ustc.edu.cn/anaconda/pkgs/main/
conda config --add channels https://mirrors.ustc.edu.cn/anaconda/cloud/conda-forge/
conda config --add channels https://mirrors.ustc.edu.cn/anaconda/cloud/bioconda/
conda config --set show_channel_urls yes
第四步:使用miniconda搜索 安装卸载软件 (以fastqc为例)
查看软件列表
conda list
# packages in environment at /root/miniconda3:
#
# Name Version Build Channel
asn1crypto 0.24.0 py36_0 defaults
ca-certificates 2018.03.07 0 defaults
certifi 2018.4.16 py36_0 defaults
cffi 1.11.5 py36h9745a5d_0 defaults
chardet 3.0.4 py36h0f667ec_1 defaults
conda 4.5.4 py36_0 defaults
conda-env 2.6.0 h36134e3_1 defaults
cryptography 2.2.2 py36h14c3975_0 defaults
idna 2.6 py36h82fb2a8_1 defaults
libedit 3.1.20170329 h6b74fdf_2 defaults
libffi 3.2.1 hd88cf55_4 defaults
libgcc-ng 7.2.0 hdf63c60_3 defaults
libstdcxx-ng 7.2.0 hdf63c60_3 defaults
ncurses 6.1 hf484d3e_0 defaults
openssl 1.0.2o h20670df_0 defaults
pip 10.0.1 py36_0 defaults
pycosat 0.6.3 py36h0a5515d_0 defaults
pycparser 2.18 py36hf9f622e_1 defaults
pyopenssl 18.0.0 py36_0 defaults
pysocks 1.6.8 py36_0 defaults
python 3.6.5 hc3d631a_2 defaults
readline 7.0 ha6073c6_4 defaults
requests 2.18.4 py36he2e5f8d_1 defaults
ruamel_yaml 0.15.37 py36h14c3975_2 defaults
setuptools 39.2.0 py36_0 defaults
six 1.11.0 py36h372c433_1 defaults
sqlite 3.23.1 he433501_0 defaults
tk 8.6.7 hc745277_3 defaults
urllib3 1.22 py36hbe7ace6_0 defaults
wheel 0.31.1 py36_0 defaults
xz 5.2.4 h14c3975_4 defaults
yaml 0.1.7 had09818_2 defaults
zlib 1.2.11 ha838bed_2 defaults
搜索fastqc软件
conda search fastqc
Loading channels: done
# Name Version Build Channel
fastqc 0.10.1 0 anaconda/cloud/bioconda
fastqc 0.10.1 1 anaconda/cloud/bioconda
fastqc 0.11.2 1 anaconda/cloud/bioconda
fastqc 0.11.2 pl5.22.0_0 anaconda/cloud/bioconda
fastqc 0.11.3 0 anaconda/cloud/bioconda
fastqc 0.11.3 1 anaconda/cloud/bioconda
fastqc 0.11.4 0 anaconda/cloud/bioconda
fastqc 0.11.4 1 anaconda/cloud/bioconda
fastqc 0.11.4 2 anaconda/cloud/bioconda
fastqc 0.11.5 1 anaconda/cloud/bioconda
fastqc 0.11.5 pl5.22.0_2 anaconda/cloud/bioconda
fastqc 0.11.5 pl5.22.0_3 anaconda/cloud/bioconda
fastqc 0.11.6 2 anaconda/cloud/bioconda
fastqc 0.11.6 pl5.22.0_0 anaconda/cloud/bioconda
fastqc 0.11.6 pl5.22.0_1 anaconda/cloud/bioconda
fastqc 0.11.7 4 anaconda/cloud/bioconda
fastqc 0.11.7 pl5.22.0_0 anaconda/cloud/bioconda
fastqc 0.11.7 pl5.22.0_2 anaconda/cloud/bioconda
安装
conda install fastqc -y
conda install fastqc=0.11.7 -y
Preparing transaction: done
Verifying transaction: done
Executing transaction: done
卸载
conda remove fastqc -y
The following packages will be REMOVED:
fastqc: 0.11.7-pl5.22.0_0 https://mirrors.ustc.edu.cn/anaconda/cloud/bioconda
Preparing transaction: done
Verifying transaction: done
Executing transaction: done
第五步:建立conda环境,环境中软件的安装和卸载
查看环境
conda info --envs
# conda environments:
#
base * /root/miniconda3
建立一个叫rnaseq的conda环境,然后指定python版本是3,安装软件fastqc、trimmomatic
conda create -n rna-seq python=3 fastqc trimmomatic -ycdca
再次查看环境
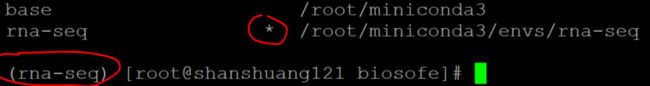
conda info --envs
# conda environments:
#
base * /root/miniconda3
rna-seq /root/miniconda3/envs/rna-seq
激活新的conda环境
source activate rna-seq
改变
输入fastqc看一看
fastqc
Exception in thread "main" java.awt.HeadlessException:
No X11 DISPLAY variable was set, but this program performed an operation which requires it.
at java.awt.GraphicsEnvironment.checkHeadless(GraphicsEnvironment.java:204)
at java.awt.Window.(Window.java:536)
at java.awt.Frame.(Frame.java:420)
at java.awt.Frame.(Frame.java:385)
at javax.swing.JFrame.(JFrame.java:189)
at uk.ac.babraham.FastQC.FastQCApplication.(FastQCApplication.java:63)
at uk.ac.babraham.FastQC.FastQCApplication.main(FastQCApplication.java:332)
卸载一个环境中的软件
conda remove -n rna-seq fastqc -y
卸载一个环境中全部软件
conda remove -n rna-seq -all
usage: conda [-h] [-V] command ...
conda: error: unrecognized arguments: -all
出错了,不知道为什么?是因为就剩一个软件了吗