数据挖掘---营销推广预测响应
数据集及源码:https://download.csdn.net/download/qq_42363032/12645777
文章目录
-
- 预测
-
- 1.读取数据
- 2.提取X和Y
- 3.连续型变量特征重要性筛选(相关系数)
-
- 将相关系数小于0.1的删除(弱相关)
- 筛选之后的数值型特征
- 4.离散型变量特征筛选(方差分析)
-
- 筛选之后的离散型特征
- 5.特征重新拼接
-
- 看一下连续型特征的基本统计量(EDA探索性分析)
- 连续型变量处理
-
- 对数处理
- 看一下离散型特征的基本统计量(EDA探索性分析)
- 离散型变量处理
- 6.前向选择法筛选变量
- 7.建立线性回归模型
- 回归
-
- 1.读入数据
-
- 一个经验的做法,对特征去重,根据去重后的个数是否大于10,来判断特征是数值型还是离散型
- 2.特征筛选(卡方检验)
-
- 以DemGender列为例
- 以StatusCat96NK列为例
- 3.通过WOE、IV看一下特征筛选
-
- 底层实现
- 调包实现
-
- 根据IV值筛选离散变量
- 根据IV值筛选连续型变量
- 4.EDA探索性分析
-
- 消除异常值
-
- 1.均值加减三倍标准差,盖帽法替换
- 2.计算百分数
- 5.针对有问题的变量进行修改
-
- 连续型变量修改
- 离散型变量修改
- 6.通过随机森林对变量的重要性进行筛选
- 7.变量聚类
- 8.逻辑回归建模
- 9.ROC曲线
预测
1.读取数据
做预测,所以离散列TARGET_B删除
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
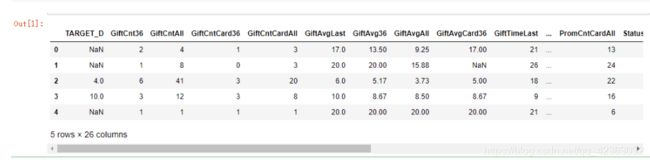
model_data = pd.read_csv("donations2.csv").drop(["ID","TARGET_B"],1) # 做预测,离散值TARGET_B删除
model_data.head()
2.提取X和Y
做预测,Y是连续型
特征有好多列,既有数值型又有离散型,这里把数值型取出为一组,离散型取出为一组,分别用相关系数和方差分析做特征筛选
print(model_data.dtypes) # 看一下数据类型
y = ["TARGET_D"]
# 提取X分为数值类型和非数值类型
var_c = ["GiftCnt36","GiftCntAll","GiftCntCard36","GiftCntCardAll","GiftTimeLast",
"GiftTimeFirst","PromCnt12","PromCnt36","PromCntAll","PromCntCard12",
"PromCntCard36","PromCntCardAll","StatusCatStarAll","DemAge",
"DemMedHomeValue","DemPctVeterans","DemMedIncome","GiftAvgLast",
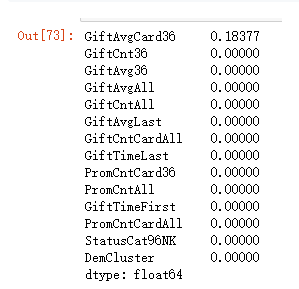
"GiftAvg36","GiftAvgAll","GiftAvgCard36"]
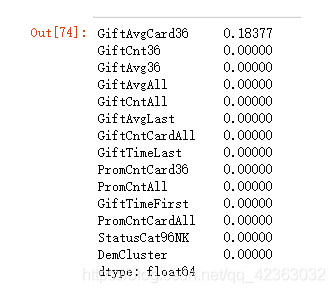
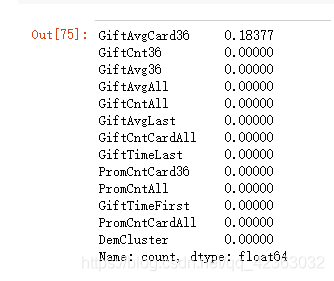
var_d = ['DemGender', 'StatusCat96NK', 'DemCluster', 'DemHomeOwner']
X = model_data[var_c + var_d]
Y = model_data[y]
3.连续型变量特征重要性筛选(相关系数)
X和Y都是数值型用相关系数
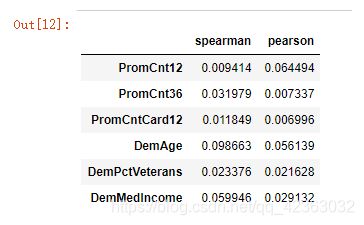
corr_s = abs(model_data[y + var_c].corr(method = 'spearman')) # spearman:斯皮尔曼相关系数
# print(corr_s['TARGET_D'])
corr_s = pd.DataFrame(corr_s.iloc[0,:]) # 第一列是TARGET_D
corr_p = abs(model_data[y + var_c].corr(method = 'pearson')) # pearson:皮尔逊相关系数
corr_p = pd.DataFrame(corr_p.iloc[0,:])
corr_sp = pd.concat([corr_s,corr_p], axis = 1)
corr_sp.columns = ['spearman','pearson']
corr_sp
将相关系数小于0.1的删除(弱相关)
corr_sp[(corr_sp['spearman'] <= 0.1) & (corr_sp['pearson'] <= 0.1)]
筛选之后的数值型特征
var_c_s = set(var_c) - set(['PromCnt12','PromCnt36',
'PromCntCard12','DemAge',
'DemPctVeterans','DemMedIncome']) # 集合的差集运算
var_c_s = list(var_c_s)
var_c_s
4.离散型变量特征筛选(方差分析)
X和Y既有离散型又有数值型用方差分析
# 当X和Y既有离散值又有连续值,通过方差分析看二者之间关系
# P值大于0.05的,需要删除,这些特征都是没有影响的没有关系的
import statsmodels.stats.anova as anova
from statsmodels.formula.api import ols
for i in var_d:
formula = "TARGET_D ~ " + str(i)
print(anova.anova_lm(ols(formula,data = model_data[var_d+['TARGET_D']]).fit())) # 方差分析是线性方程的特例
P值大于0.05的需要删除,说明此特征对Y没有影响没有关系
筛选之后的离散型特征
var_d_s = list(set(var_d) - set(["DemHomeOwner"]))
var_d_s
5.特征重新拼接
X = model_data[var_c_s + var_d_s].copy()
Y = model_data[y].copy()
model_data[var_c_s+var_d_s].head()
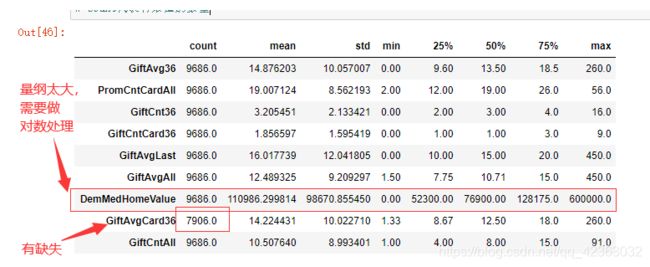
看一下连续型特征的基本统计量(EDA探索性分析)
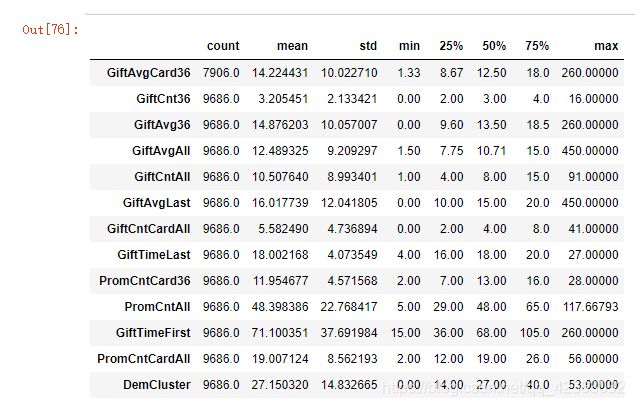
X[var_c_s].describe().T # 看一下数值型特征的描述性信息,基本的统计量
# count代表有效值的数量
abs((X[var_c_s].mode().iloc[0,] - X[var_c_s].median()) /
(X[var_c_s].quantile(0.75) - X[var_c_s].quantile(0.25)))
# (众数 - 中位数)/(上四分位数 - 下四分位数)【极差】
# 如果值大于0.8或者大于1,说明这个数稍微有点偏了,有可能存在异常值
看一下直方图和偏度值
X["PromCntAll"].hist()
X["DemMedHomeValue"].hist()
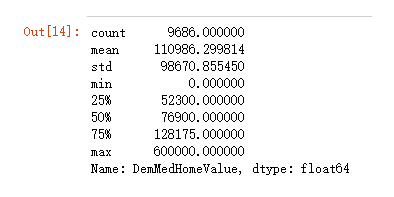
X["DemMedHomeValue"].describe().T
X['DemMedHomeValue'].replace(0, np.nan, inplace = True)
说明PromCntAll和DemMedHomeValue需要做对数处理
连续型变量处理
1 - (X.describe().T["count"]) / len(X)
GiftAvgCard36_fill = X["GiftAvgCard36"].median() # 中位数
DemMedHomeValue_fill = X["DemMedHomeValue"].median()
X["GiftAvgCard36"].fillna(GiftAvgCard36_fill,inplace = True) # 用中位数进行缺失值填充
X["DemMedHomeValue"].fillna(DemMedHomeValue_fill,inplace = True)
skew_var_x = {
}
for i in var_c_s:
skew_var_x[i]=abs(X[i].skew()) # 看一下峰值
skew = pd.Series(skew_var_x).sort_values(ascending=False)
skew
var_x_ln = skew[skew >= 1].index #返回大于等于1的索引
var_x_ln
对数处理
# 对数处理
for i in var_x_ln:
if min(X[i]) <= 0:
X[i] = np.log(X[i] + abs(min(X[i])) + 0.01)
else:
X[i] = np.log(X[i])
# 看一下峰值
skew_var_x = {
}
for i in var_c_s:
skew_var_x[i]=abs(X[i].skew())
skew = pd.Series(skew_var_x).sort_values(ascending=False)
skew
看一下离散型特征的基本统计量(EDA探索性分析)
对于离散型的变量没有办法算统计量,一般情况做一下频数分布的计算
类别数一般6-8类就是比较不错的
X["DemGender"].value_counts() # 这个特征只有3类,还可以
X["StatusCat96NK"].value_counts() #有的水平数量太少,需要合并,类别少的归成一类
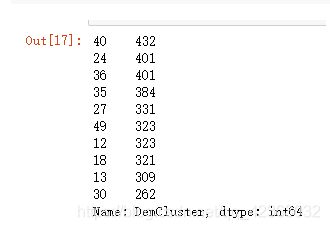
X["DemCluster"].value_counts()[:10] # 这个类别太多,好像50多个,也需要合并,比如合并到8-10类
离散型变量处理
对于DemCluster
# as_index 的默认值为True, 对于聚合输出,返回以组标签作为索引的对象。仅与DataFrame输入相关。as_index = False实际上是“SQL风格”的分组输出
DemC_group = model_data[['DemCluster','TARGET_D']].groupby('DemCluster',as_index = False)
DemC_C= DemC_group['TARGET_D'].agg({
'mean' : 'mean','count':'count',"median":"median"}).sort_values(["median","mean"])
DemC_C["count_cumsum"] = DemC_C["count"].cumsum()
DemC_C["new_DemCluster"] = DemC_C["count_cumsum"].apply(lambda x:x//(len(model_data)/10)+1)
DemC_C["new_DemCluster"] = DemC_C["new_DemCluster"].astype(int)
DemC_C
len(model_data)
DemCluster_new_class = DemC_C[["DemCluster","new_DemCluster"]].set_index("DemCluster")
X["DemCluster"] = X["DemCluster"].map(DemCluster_new_class.to_dict()['new_DemCluster'])
new_DemGender = {
"F":1,"M":2,"U":3}
X['DemGender'] = X['DemGender'].map(new_DemGender)
对于StatusCat96NK
StatusCat96NK_group = model_data[['StatusCat96NK','TARGET_D']].groupby('StatusCat96NK', as_index = False)
StatusCat96NK_class = StatusCat96NK_group['TARGET_D'].agg({
'mean' : 'mean', 'count':'count',"median":"median"}).sort_values(["median","mean"])
StatusCat96NK_class
new_StatusCat96NK = {
"S":1,"A":2,"E":2,"N":2,"F":2,"L":2}
X['StatusCat96NK'] = X['StatusCat96NK'].map(new_StatusCat96NK)
X.head()
6.前向选择法筛选变量
import statsmodels.formula.api as smf
def forward_selected(data, response):
remaining = set(data.columns)
remaining.remove(response)
selected = []
current_score, best_new_score = 0.0, 0.0
while remaining and current_score == best_new_score:
scores_with_candidates = []
for candidate in remaining:
formula = "{} ~ {} + 1".format(response,
' + '.join(selected + [candidate]))
score = smf.ols(formula, data).fit().rsquared_adj
scores_with_candidates.append((score, candidate))
scores_with_candidates.sort()
best_new_score, best_candidate = scores_with_candidates.pop()
if current_score < best_new_score:
remaining.remove(best_candidate)
selected.append(best_candidate)
current_score = best_new_score
formula = "{} ~ {} + 1".format(response,
' + '.join(selected))
model = smf.ols(formula, data).fit()
return selected
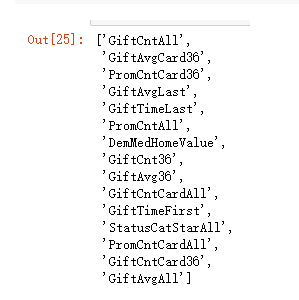
XX=pd.concat([X[var_c_s+var_d_s],Y],axis = 1)
final_var = forward_selected(XX,'TARGET_D')
var_c_s = list(set(var_c_s)&set(final_var))
var_c_s
var_d_s = list(set(var_d_s)&set(final_var))
var_d_s
model_final = pd.concat([X[var_d_s + var_c_s],Y], axis = 1)
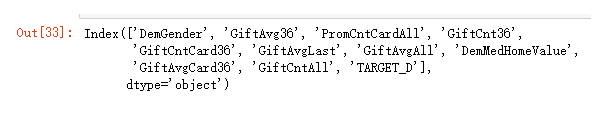
model_final.columns
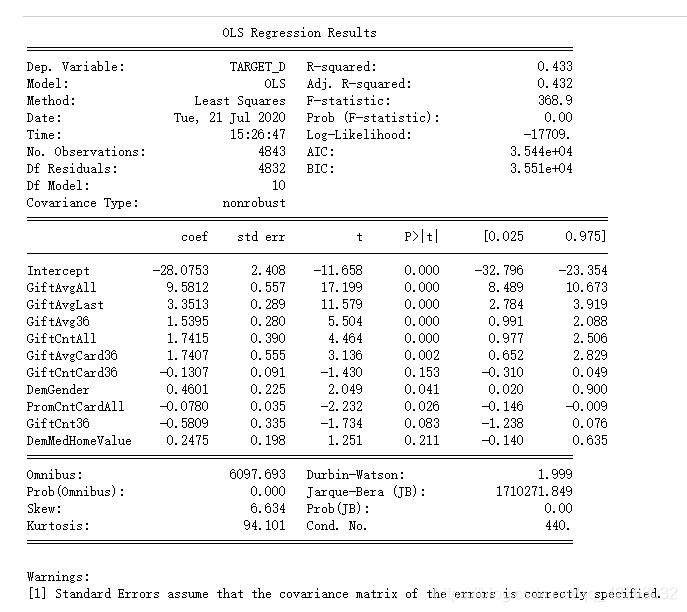
7.建立线性回归模型
import statsmodels.api as sm
from statsmodels.formula.api import ols
X = model_final.iloc[:,:-1]
Y = model_final.iloc[:,-1]
formula = 'TARGET_D ~ ' + '+'.join(final_var)
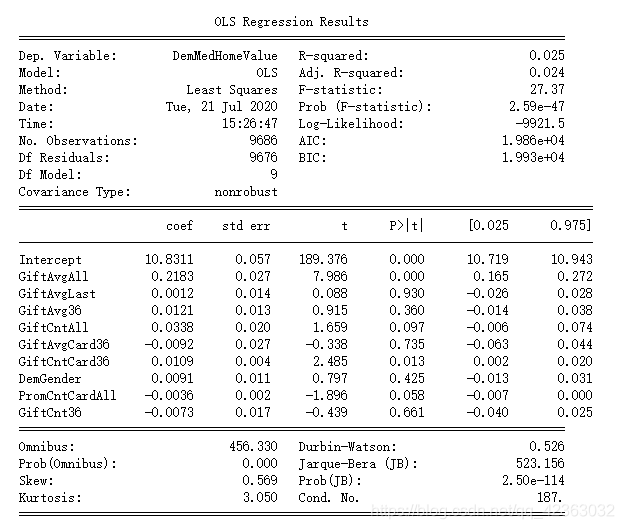
donation_model = ols(formula,model_final).fit()
print(donation_model.summary())
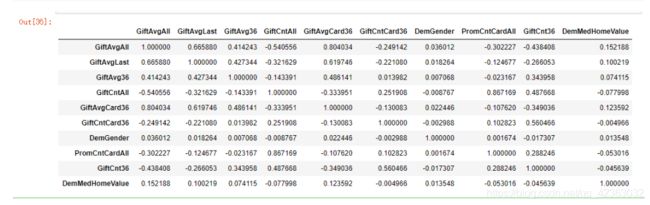
model_final[final_var].corr()
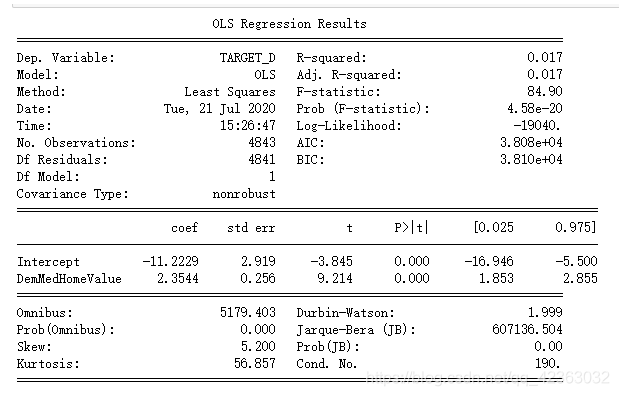
用DemMedHomeValue做线性回归模型看一下结果
formula = 'TARGET_D ~ DemMedHomeValue'
model = ols(formula,model_final).fit()
print(model.summary())
看一下特征共线性
final_varN = ['GiftAvgAll',
'GiftAvgLast',
'GiftAvg36',
'GiftCntAll',
'GiftAvgCard36',
'GiftCntCard36',
'DemGender',
'PromCntCardAll',
'GiftCnt36']
formula = 'DemMedHomeValue ~ ' + '+'.join(final_varN)
model = ols(formula,model_final).fit()
print(model.summary())
formula = 'TARGET_D ~ ' + '+'.join(final_varN)
model = ols(formula,model_final).fit()
print(model.summary())
回归
1.读入数据
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# ID TARGET_D 删除这两列
model_data=pd.read_csv("donations2.csv").drop(["ID","TARGET_D"],1)
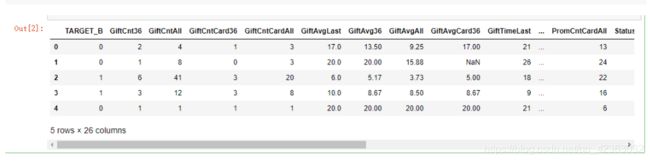
model_data.head()
一个经验的做法,对特征去重,根据去重后的个数是否大于10,来判断特征是数值型还是离散型
比如,判断GiftCntAll这一列是数值型还是离散型
len(model_data['GiftCntAll'].drop_duplicates()) # 69,大于10,说明是数值型
model_data.dtypes
y = 'TARGET_B'
# 数值类型
var_c = ["GiftCnt36","GiftCntAll","GiftCntCard36",
"GiftCntCardAll","GiftTimeLast","GiftTimeFirst",
"PromCnt12","PromCnt36","PromCntAll",
"PromCntCard12","PromCntCard36","PromCntCardAll",
"StatusCatStarAll","DemAge","DemMedHomeValue",
"DemPctVeterans","DemMedIncome","GiftAvgLast",
"GiftAvg36","GiftAvgAll","GiftAvgCard36"]
# 类别类型
var_d = ['StatusCat96NK', 'DemHomeOwner', 'DemGender', 'DemCluster']
X = model_data[var_c + var_d].copy()
Y = model_data[y].copy()
2.特征筛选(卡方检验)
当Y和X都是离散型时,用卡方检验做特征筛选
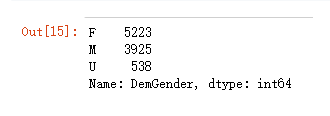
以DemGender列为例
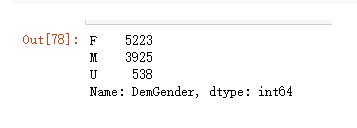
先看一下频数统计
model_data['DemGender'].value_counts()
看一下列联交叉表
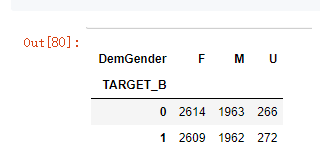
# 非参数统计------------------列联交叉表
cross = pd.crosstab(model_data['TARGET_B'],model_data['DemGender'])
cross
用这个列联交叉表做这个特征的卡方检验
from scipy import stats
from pprint import pprint
pprint(stats.chi2_contingency(cross)) # 卡方值、概率P值、自由度
DemGender列的概率P值是大于0.05的,所以是需要删除的,它对结果没有影响
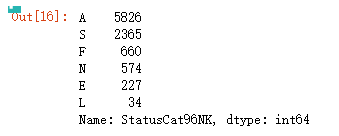
以StatusCat96NK列为例
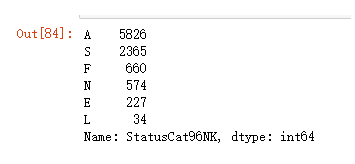
频数统计
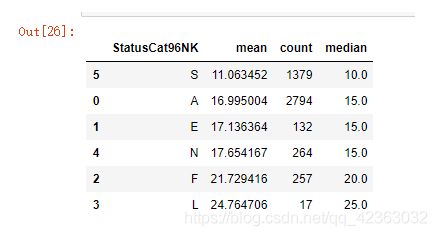
model_data['StatusCat96NK'].value_counts()
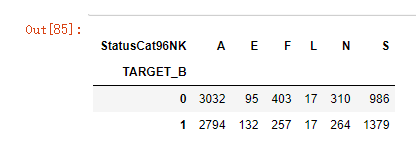
列联交叉
# 非参数统计------------------列联交叉表
cross = pd.crosstab(model_data['TARGET_B'],model_data['StatusCat96NK'])
cross
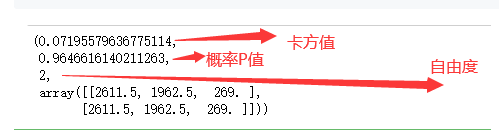
卡方检验
from scipy import stats
from pprint import pprint
pprint(stats.chi2_contingency(cross)) # 卡方值、概率P值、自由度
3.通过WOE、IV看一下特征筛选
底层实现
def CalcWOE(df, col, target):
'''
:param df: 包含需要计算WOE的变量和目标变量
:param col: 需要计算WOE、IV的变量,必须是分箱后的变量,或者不需要分箱的类别型变量,也就是离散型
:param target: 目标变量,0、1表示好、坏
:return: 返回WOE和IV
'''
total = df.groupby([col])[target].count() # 对col列根据类别标签做一下分组统计,求的是每个col标签的样本总数
total = pd.DataFrame({
'total': total}) # 下面需要做表关联,转换成数据框
bad = df.groupby([col])[target].sum() # 每个col标签的坏样本总数(1的)
bad = pd.DataFrame({
'bad': bad})
regroup = total.merge(bad, left_index=True, right_index=True, how='left')
regroup.reset_index(level=0, inplace=True) # 索引重置
N = sum(regroup['total'])
B = sum(regroup['bad'])
regroup['good'] = regroup['total'] - regroup['bad'] # 好样本数
G = N - B
regroup['bad_pcnt'] = regroup['bad'].map(lambda x: x*1.0/B) # 坏样本率
regroup['good_pcnt'] = regroup['good'].map(lambda x: x * 1.0 / G) # 好样本率
regroup['WOE'] = regroup.apply(lambda x: np.log(x.good_pcnt*1.0/x.bad_pcnt),axis = 1)
WOE_dict = regroup[[col,'WOE']].set_index(col).to_dict(orient='index')
for k, v in WOE_dict.items():
WOE_dict[k] = v['WOE']
IV = regroup.apply(lambda x: (x.good_pcnt-x.bad_pcnt)*np.log(x.good_pcnt*1.0/x.bad_pcnt),axis = 1)
IV = sum(IV)
return {
"WOE": WOE_dict, 'IV':IV}
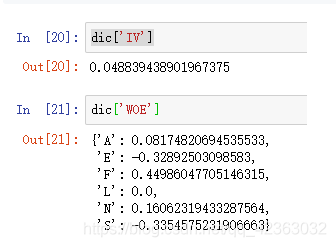
dic = CalcWOE(model_data, 'StatusCat96NK', 'TARGET_B')
dic['IV']
dic['WOE']
一般来说某个特征的IV值大于0.02,越大越好,越大说明特征越重要,可以理解随机森林的特征重要度等价
WOE入摸
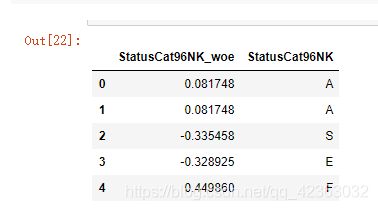
model_data['StatusCat96NK_woe'] = model_data['StatusCat96NK'].map(dic['WOE'])
model_data[['StatusCat96NK_woe','StatusCat96NK']].head()
调包实现
from woe import WoE
iv_d = {
}
for i in var_d:
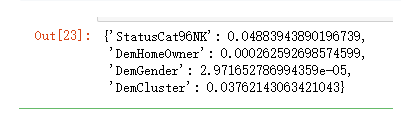
iv_d[i] = WoE(v_type='d').fit(X[i].copy(), Y.copy()).iv
iv_d
根据IV值筛选离散变量
from woe import WoE
iv_d = {
}
for i in var_d:
iv_d[i] = WoE(v_type='d').fit(X[i].copy(), Y.copy()).iv # 参数是离散型 v_type=‘d’
iv_d
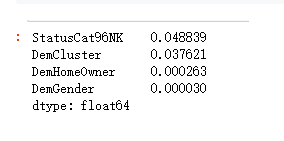
pd.Series(iv_d).sort_values(ascending = False)
var_d_s = ['StatusCat96NK', 'DemCluster']
根据IV值筛选连续型变量
iv_c = {
}
for i in var_c:
iv_c[i] = WoE(v_type='c',t_type='b',qnt_num=3).fit(X[i],Y).iv # 参数是数值型 v_type=‘c’
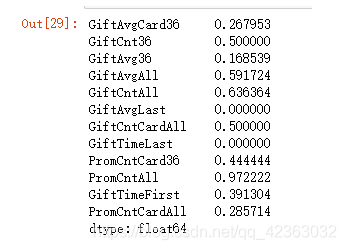
sort_iv_c = pd.Series(iv_c).sort_values(ascending=False)
sort_iv_c
保留iv值大于0.02的
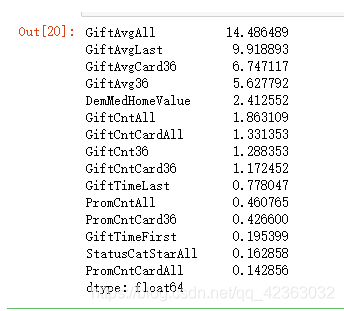
var_c_s = list(sort_iv_c[sort_iv_c > 0.02].index)
var_c_s
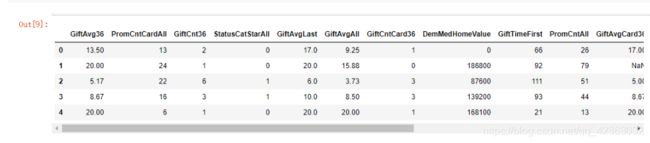
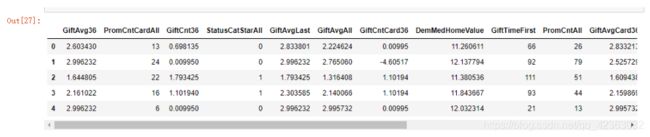
X = model_data[var_c_s + var_d_s].copy()
Y = model_data[y].copy()
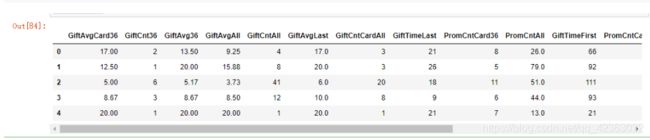
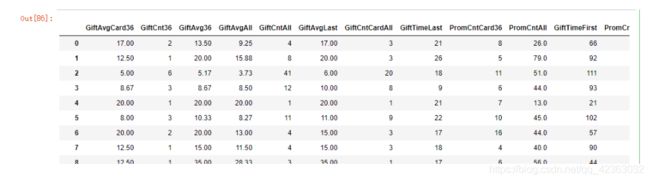
X[var_c_s].describe().T
![]()
4.EDA探索性分析
就是描述性统计加可视化
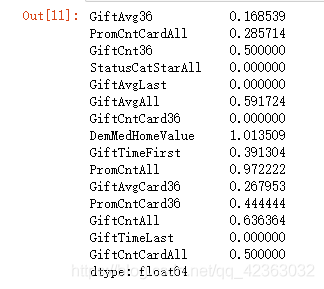
(众数 - 中位数)/(分位数极差)
经验来看,一般这个值大于0.8,说明它有偏,画一下直方图看看
abs((X[var_c_s].mode().iloc[0,] - X[var_c_s].median()) /
(X[var_c_s].quantile(0.75) - X[var_c_s].quantile(0.25)))
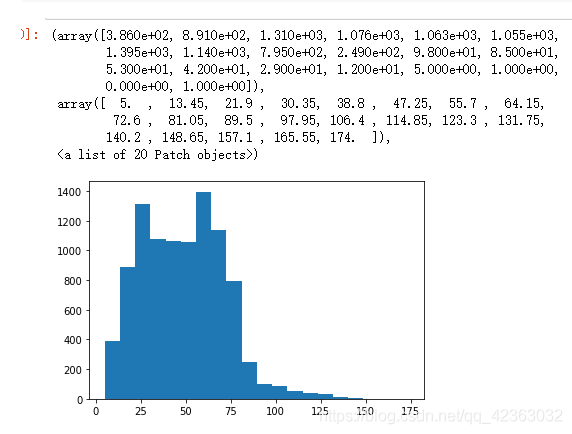
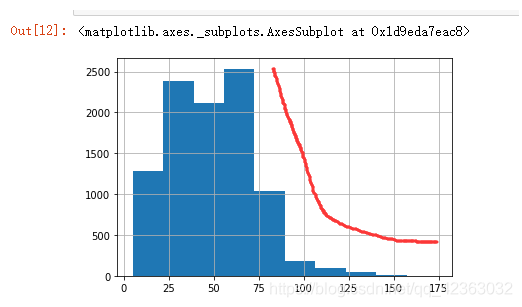
plt.hist(X["PromCntAll"], bins=20)
柱形图右面偏离是异常值
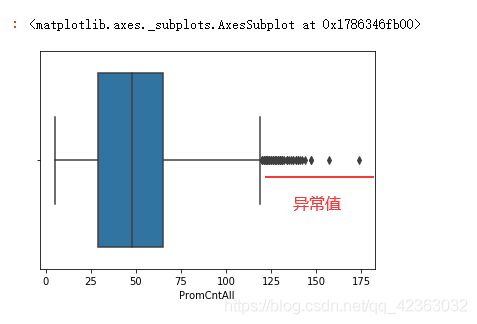
# 箱线图
# 上边缘的计算方法是 上四分位数+1.5倍极差
import seaborn as sns
sns.boxplot(X["PromCntAll"])
消除异常值
1.均值加减三倍标准差,盖帽法替换
prc_mean = X["PromCntAll"].mean()
prc_std = X["PromCntAll"].std()
high = prc_mean + 3*prc_std
low = prc_mean - 3*prc_std
def s(x):
if x > high:
return high
elif x < low:
return low
else:
return x
2.计算百分数
# 计算百分位数
p1 = X["PromCntAll"].quantile(0.01)
p99 = X["PromCntAll"].quantile(0.99)
print(p1)
print(p99)
用均值加减三倍标准差消除异常值
X["PromCntAll"] = X["PromCntAll"].map(s)
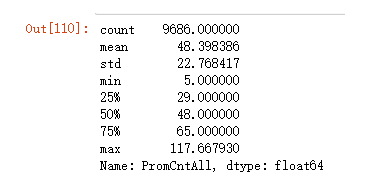
X["PromCntAll"].describe()
X["PromCntAll"][X["PromCntAll"] > low][X["PromCntAll"] < high].describe()
5.针对有问题的变量进行修改
1 - X.count()/ len(X)
X.isnull().sum()/len(X)
1 - (X.describe().T["count"]) / len(X)
X.describe().T
连续型变量修改
fill_GiftAvgCard36 = X.GiftAvgCard36.median()
X.GiftAvgCard36.fillna(value=fill_GiftAvgCard36, inplace=True)
X.GiftAvgCard36.fillna(method='ffill', inplace=True)
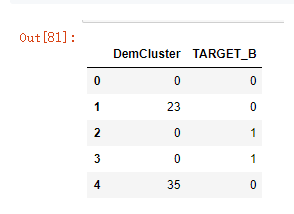
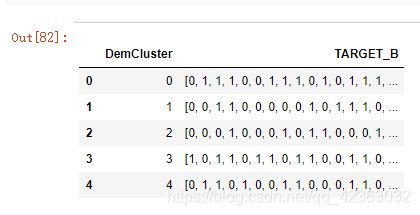
model_data[['DemCluster','TARGET_B']].head()
model_data[['DemCluster','TARGET_B']].groupby('DemCluster',as_index = False).agg(list).head()
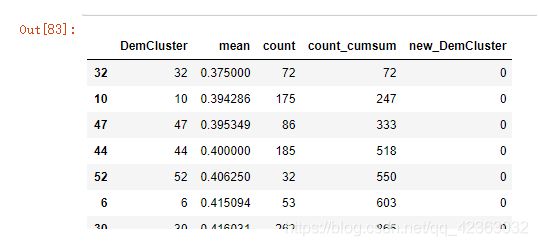
离散型变量修改
# 统计每个水平的对应目标变量的均值,和每个水平数量
DemCluster_grp = model_data[['DemCluster',
'TARGET_B']].groupby('DemCluster',as_index = False)
DemC_C = DemCluster_grp['TARGET_B'].agg({
'mean' : 'mean',
'count':'count'}).sort_values("mean")
# 将这些类别尽量以人数大致均等的方式以响应率为序归结为10个大类
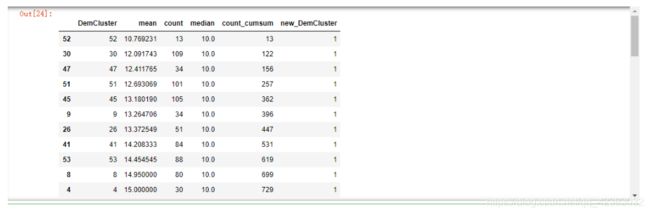
DemC_C["count_cumsum"] = DemC_C["count"].cumsum()
DemC_C["new_DemCluster"] = DemC_C["count_cumsum"].apply(lambda x: x//(len(model_data)/10)).astype(int)
DemC_C
X_rep = X.copy()
for i in var_d_s:
X_rep[i+"_woe"] = WoE(v_type='d').fit_transform(X_rep[i],Y)
X_rep.head()
StatusCat96NK_woe = X_rep[["StatusCat96NK","StatusCat96NK_woe"]].drop_duplicates().set_index("StatusCat96NK").to_dict()
StatusCat96NK_woe
StatusCat96NK_woe = X_rep[["StatusCat96NK","StatusCat96NK_woe"]].drop_duplicates().set_index("StatusCat96NK").to_dict()
DemCluster_woe = X_rep[["DemCluster","DemCluster_woe"]].drop_duplicates().set_index("DemCluster").to_dict()
del X_rep["StatusCat96NK"]
del X_rep["DemCluster"]
X_rep.rename(columns={
"StatusCat96NK_woe":"StatusCat96NK","DemCluster_woe":"DemCluster"},inplace=True)
X_rep
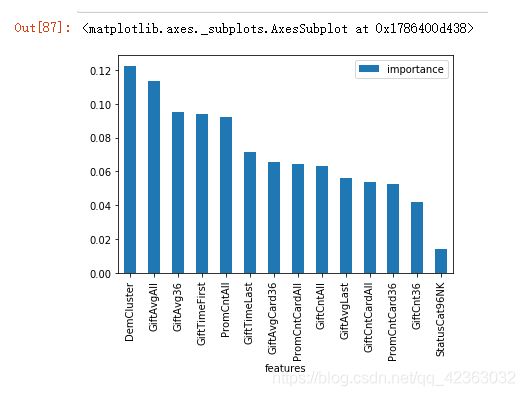
6.通过随机森林对变量的重要性进行筛选
import sklearn.ensemble as ensemble
rfc = ensemble.RandomForestClassifier(criterion='entropy', n_estimators=3, max_features=0.5, min_samples_split=5)
rfc_model = rfc.fit(X_rep, Y)
rfc_model.feature_importances_
rfc_fi = pd.DataFrame()
rfc_fi["features"] = list(X.columns)
rfc_fi["importance"] = list(rfc_model.feature_importances_)
rfc_fi=rfc_fi.set_index("features",drop=True)
var_sort = rfc_fi.sort_values(by="importance",ascending=False)
var_sort.plot(kind="bar")
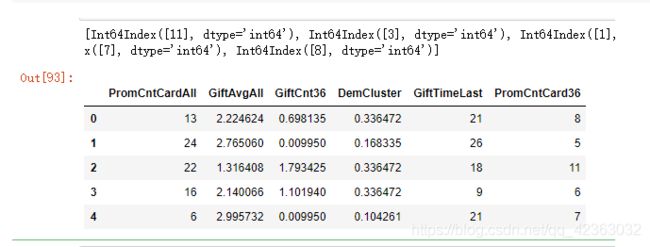
7.变量聚类
每一类选一个具有代表性的
权重向量稀疏解的一个计算
from VarSelec import Var_Select,Var_Select_auto
X_rep_reduc = Var_Select_auto(X_rep)
X_rep_reduc.head()
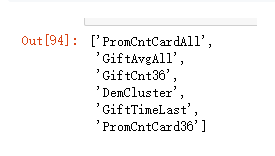
list(X_rep_reduc.columns)
8.逻辑回归建模
import sklearn.model_selection as model_selection
ml_data = model_selection.train_test_split(X_rep_reduc, Y, test_size=0.3, random_state=0)
train_data, test_data, train_target, test_target = ml_data
from sklearn import preprocessing
min_max_scaler = preprocessing.MinMaxScaler()
train_data = min_max_scaler.fit_transform(train_data)
test_data = min_max_scaler.fit_transform(test_data)
train_data
import sklearn.linear_model as linear_model
logistic_model = linear_model.LogisticRegression(class_weight = None,
dual = False,
fit_intercept = True,
intercept_scaling = 1,
penalty = 'l2',
random_state = None,
tol = 0.001)
from sklearn.model_selection import ParameterGrid, GridSearchCV
C = np.logspace(-3,0,20,base=10)
param_grid = {
'C': C}
clf_cv = GridSearchCV(estimator=logistic_model,
param_grid=param_grid,
cv=5,
scoring='roc_auc')
clf_cv.fit(train_data, train_target)
import statsmodels.api as sm
import statsmodels.formula.api as smf
model=X_rep_reduc.join(train_target)
formula = "TARGET_B ~ " + "+".join(X_rep_reduc)
lg_m = smf.glm(formula=formula, data=model, family=sm.families.Binomial(sm.families.links.logit)).fit()
lg_m.summary()
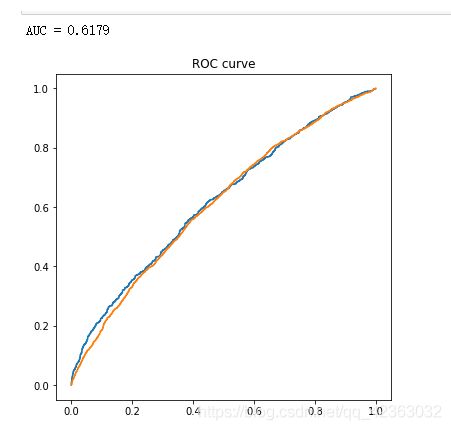
9.ROC曲线
test_est = logistic_model.predict(test_data)
train_est = logistic_model.predict(train_data)
test_est_p = logistic_model.predict_proba(test_data)[:,1]
train_est_p = logistic_model.predict_proba(train_data)[:,1]
import sklearn.metrics as metrics
fpr_test, tpr_test, th_test = metrics.roc_curve(test_target, test_est_p)
fpr_train, tpr_train, th_train = metrics.roc_curve(train_target, train_est_p)
plt.figure(figsize=[6,6])
plt.plot(fpr_test, tpr_test)
plt.plot(fpr_train, tpr_train)
plt.title('ROC curve')
print('AUC = %6.4f' %metrics.auc(fpr_test, tpr_test))



![[外链图片转存失败,源站可能有防盗链机制,建议将图片保存下来直接上传(img-aHBsFoPI-1595762495884)(FED7B002F7A44961B1A8A2F2F2FD03A3)]](http://img.e-com-net.com/image/info8/3f8de38d20da41548991fa479545828e.png)




![[外链图片转存失败,源站可能有防盗链机制,建议将图片保存下来直接上传(img-GfHMOaGG-1595762495908)(495195815EDC4B91A7ED497744A85A04)]](http://img.e-com-net.com/image/info8/3f6f5db506a0404e891e75c37562b250.png)

![[外链图片转存失败,源站可能有防盗链机制,建议将图片保存下来直接上传(img-6pemw6oW-1595762495913)(174E0706B9CF43C0BE835FABCDE40F84)]](http://img.e-com-net.com/image/info8/08b63dfad0c74f4f922ee79f79b3902a.png)