> library(qqman)
For example usage please run: vignette('qqman')
Citation appreciated but not required:
Turner, S.D. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. biorXiv DOI: 10.1101/005165 (2014).
> gwasResults
SNP CHR BP P
1 rs1 1 1 0.9148060435
2 rs2 1 2 0.9370754133
3 rs3 1 3 0.2861395348
4 rs4 1 4 0.8304476261
5 rs5 1 5 0.6417455189
6 rs6 1 6 0.5190959491
#第一列SNP的ID,第二列是染色体编号,第三列是SNP在染色体上的坐标(模拟数据) ,第四列是p-value#
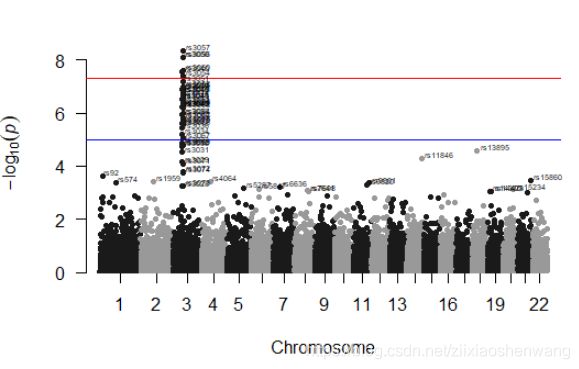
> manhattan(gwasResults)#默认无颜色,阈值线是5和7多一点#
#3号染色体上的点可能与性状相关#
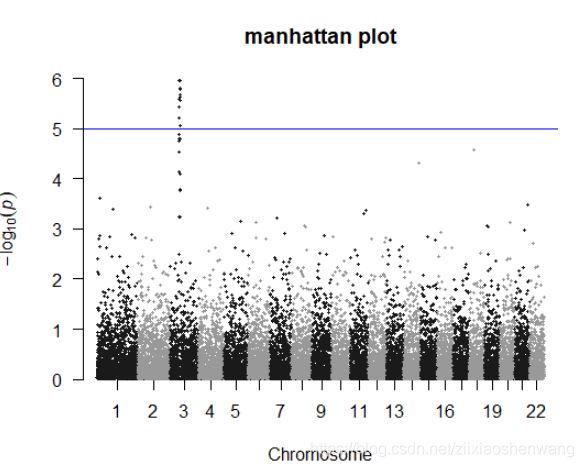
> manhattan(gwasResults,main ="manhattan plot",ylim=c(0,6))

> manhattan(gwasResults,main ="manhattan plot",ylim=c(0,6),cex = 0.6)
#cex缩放点的大小#
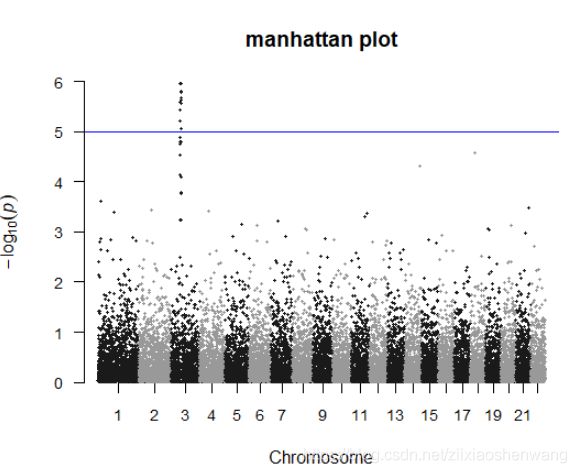
> manhattan(gwasResults,main ="manhattan plot",ylim=c(0,6),cex = 0.6,cex.axis=0.9)
> manhattan(gwasResults,main ="manhattan plot",ylim=c(0,6),cex = 0.6,cex.axis=0.9,col = c("blue4","orange3"))
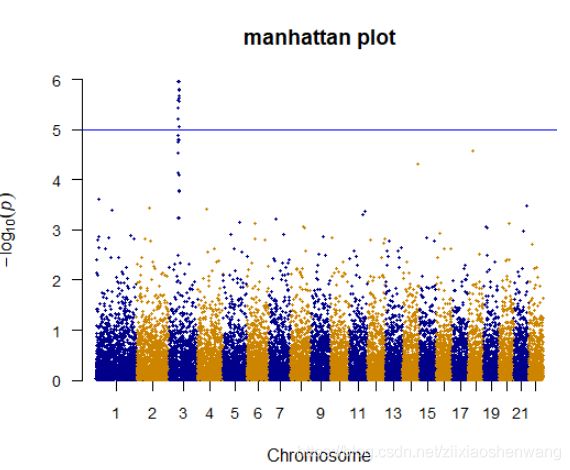
> manhattan(gwasResults,main ="manhattan plot",ylim=c(0,6),cex = 0.6,cex.axis=0.9,col = c("blue4","orange3"),suggestiveline = F)#suggestiveline提示线,默认是8#

> manhattan(gwasResults,main ="manhattan plot",ylim=c(0,6),cex = 0.6,cex.axis=0.9,col = c("blue4","orange3"),suggestiveline = F,genomewideline = F)#genomewideline线,
#一个是上限,一个是下限#
> manhattan(gwasResults,main ="manhattan plot",ylim=c(0,6),cex = 0.6,cex.axis=0.9,col = c("blue4","orange3"),suggestiveline = T,genomewideline = T)

> manhattan(gwasResults,main ="manhattan plot",ylim=c(0,6),cex = 0.6,cex.axis=0.9,col = c("blue4","orange3"),suggestiveline = 8,genomewideline = 5)

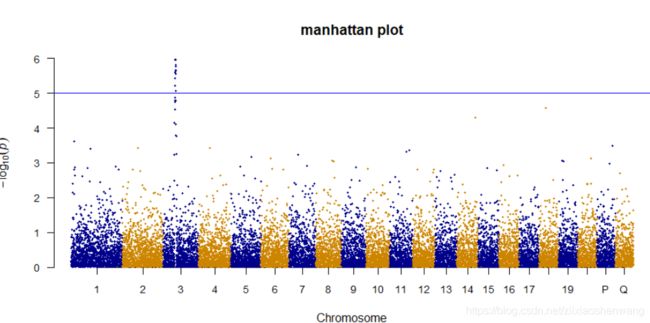
> manhattan(gwasResults,main ="manhattan plot",ylim=c(0,6),cex = 0.6,cex.axis=0.9,col = c("blue4","orange3"),chrlabs = c(1:20,"P","Q"))
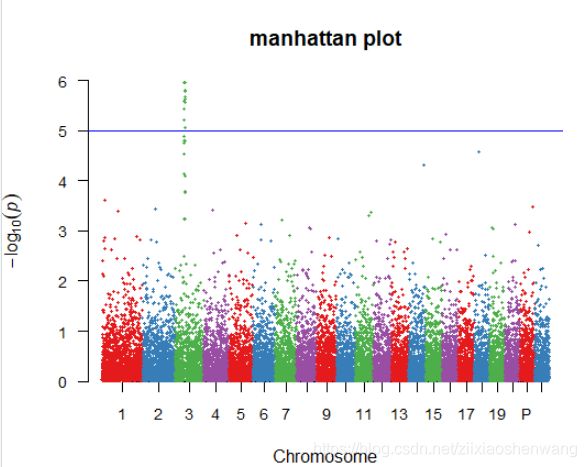
> library(RColorBrewer)
> yanse <- brewer.pal(name = "Set1",n=4)
> manhattan(gwasResults,main ="manhattan plot",ylim=c(0,6),cex = 0.6,cex.axis=0.9,col = yanse,chrlabs = c(1:20,"P","Q"))
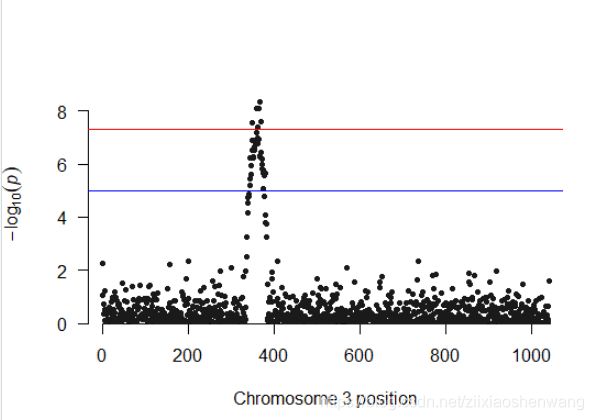
#单独绘制3好染色体#
> manhattan(subset(gwasResults,CHR==3))
> snpsOfInterest#snp位点,需要高亮的部分
[1] "rs3001" "rs3002" "rs3003" "rs3004" "rs3005" "rs3006" "rs3007" "rs3008"
[9] "rs3009" "rs3010" "rs3011" "rs3012" "rs3013" "rs3014" "rs3015" "rs3016"
[17] "rs3017" "rs3018" "rs3019" "rs3020" "rs3021" "rs3022" "rs3023" "rs3024"
[25] "rs3025" "rs3026" "rs3027" "rs3028" "rs3029" "rs3030" "rs3031" "rs3032"
[33] "rs3033" "rs3034" "rs3035" "rs3036" "rs3037" "rs3038" "rs3039" "rs3040"
[41] "rs3041" "rs3042" "rs3043" "rs3044" "rs3045" "rs3046" "rs3047" "rs3048"
[49] "rs3049" "rs3050" "rs3051" "rs3052" "rs3053" "rs3054" "rs3055" "rs3056"
[57] "rs3057" "rs3058" "rs3059" "rs3060" "rs3061" "rs3062" "rs3063" "rs3064"
[65] "rs3065" "rs3066" "rs3067" "rs3068" "rs3069" "rs3070" "rs3071" "rs3072"
[73] "rs3073" "rs3074" "rs3075" "rs3076" "rs3077" "rs3078" "rs3079" "rs3080"
[81] "rs3081" "rs3082" "rs3083" "rs3084" "rs3085" "rs3086" "rs3087" "rs3088"
[89] "rs3089" "rs3090" "rs3091" "rs3092" "rs3093" "rs3094" "rs3095" "rs3096"
[97] "rs3097" "rs3098" "rs3099" "rs3100"
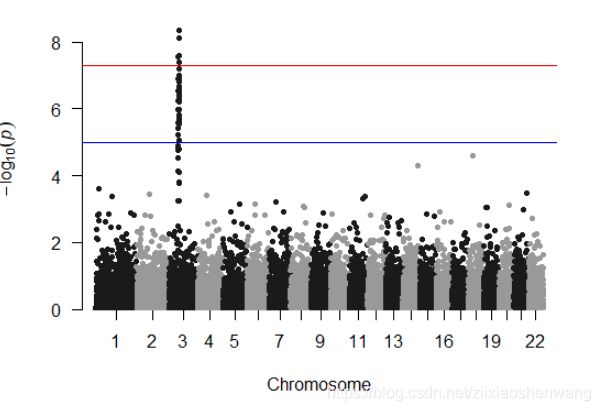
> manhattan(gwasResults,highlight=snpsOfInterest)

> manhattan(gwasResults,annotatePval = 0.001)
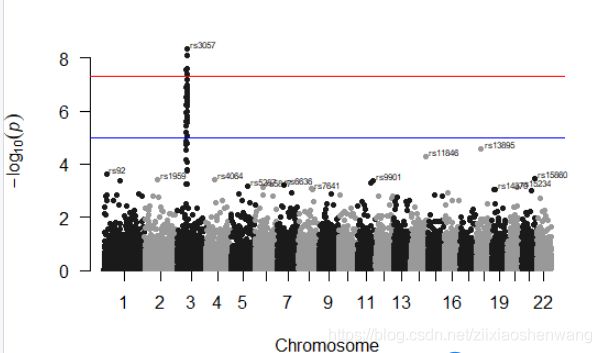
> manhattan(gwasResults,annotatePval = 0.001,annotateTop = F)#显示显著的SNP点对应的名字或者number号#