GraphSAGE 源代码 -- 分图训练
源代码下载链接:GitHub - twjiang/graphSAGE-pytorch: A PyTorch implementation of GraphSAGE. This package contains a PyTorch implementation of GraphSAGE.
1 使用数据集介绍
数据集使用cora;
图数据集,包含2708篇科学出版物, 5429条边,总共7种类别;
每篇论文至少引用一篇论文或被至少一篇论文引用(即至少有一条出边或至少有一条入边,也就是样本点之间存在联系,没有任何一个样本点与其他样本点完全没联系。如果将样本点看做图中的点,则这是一个连通的图,不存在孤立点);
在词干堵塞和去除词尾后,且文档频率小于10的所有单词都被删除后,只剩下1433个单词;
Cora 数据集中主要包含两个文件:cora.content 和 cora.cites;
cora.content内容展示:
主要包含三部分: 论文ID, 论文特征表示, 论文类别
31336 0 0 0 0 0 0 0 0 ... 0 Neural_Networks
1061127 0 0 0 0 ... 0 0 0 0 0 Rule_Learning
1106406 0 ... 0 0 0 0 0 0 0 0 0 Reinforcement_Learning
cora.cites内容展示: 两组论文编号,表示其之间有边;
35 1033
35 103482
35 103515
35 1050679
35 11039602 主函数部分 src/main.py
第一部分设置training set
import sys
import os
import torch
import argparse
import pyhocon
import random
from src.dataCenter import *
from src.utils import *
from src.models import *
parser = argparse.ArgumentParser(description='pytorch version of GraphSAGE')
parser.add_argument('--dataSet', type=str, default='cora')
parser.add_argument('--agg_func', type=str, default='MEAN')
parser.add_argument('--epochs', type=int, default=50)
parser.add_argument('--b_sz', type=int, default=20)
parser.add_argument('--seed', type=int, default=824)
parser.add_argument('--cuda', action='store_true',
help='use CUDA')
parser.add_argument('--gcn', action='store_true')
parser.add_argument('--learn_method', type=str, default='sup')
parser.add_argument('--unsup_loss', type=str, default='normal')
parser.add_argument('--max_vali_f1', type=float, default=0)
parser.add_argument('--name', type=str, default='debug')
parser.add_argument('--config', type=str, default='experiments.conf')
args = parser.parse_args()
if torch.cuda.is_available():
if not args.cuda:
print("WARNING: You have a CUDA device, so you should probably run with --cuda")
else:
device_id = torch.cuda.current_device()
print('using device', device_id, torch.cuda.get_device_name(device_id))
device = torch.device("cuda" if args.cuda else "cpu")
print('DEVICE:', device)
if __name__ == '__main__':
random.seed(args.seed)
np.random.seed(args.seed)
torch.manual_seed(args.seed)
torch.cuda.manual_seed_all(args.seed)
# load config file
# 导入training set
config = pyhocon.ConfigFactory.parse_file(args.config)读取数据:
# load data
ds = args.dataSet
dataCenter = DataCenter(config)
dataCenter.load_dataSet(ds)
# 取出节点的特征
features = torch.FloatTensor(getattr(dataCenter, ds+'_feats')).to(device)设置graphsage;
graphSage = GraphSage(config['setting.num_layers'], features.size(1), config['setting.hidden_emb_size'], features, getattr(dataCenter, ds+'_adj_lists'), device, gcn=args.gcn, agg_func=args.agg_func)
graphSage.to(device)
# 定义label的数量 7
num_labels = len(set(getattr(dataCenter, ds+'_labels')))
# graphsage输出特征后,经过分类器
classification = Classification(config['setting.hidden_emb_size'], num_labels)
classification.to(device)因为graphsage涉及到有监督和无监督,这里设置一个无监督的loss;
# 目前采用的是有监督学习模型,这里可以不看
unsupervised_loss = UnsupervisedLoss(getattr(dataCenter, ds+'_adj_lists'), getattr(dataCenter, ds+'_train'), device)
然后是训练模型;
训练了两个模型,第一个是graphsage, 第二个是分类模型;
# 判定学习类型,这里是采用有监督模型的
if args.learn_method == 'sup':
print('GraphSage with Supervised Learning')
elif args.learn_method == 'plus_unsup':
print('GraphSage with Supervised Learning plus Net Unsupervised Learning')
else:
print('GraphSage with Net Unsupervised Learning')
for epoch in range(args.epochs):
print('----------------------EPOCH %d-----------------------' % epoch)
# apply_model模型运行的函数
graphSage, classification = apply_model(dataCenter, ds, graphSage, classification, unsupervised_loss, args.b_sz, args.unsup_loss, device, args.learn_method)
if (epoch+1) % 2 == 0 and args.learn_method == 'unsup':
classification, args.max_vali_f1 = train_classification(dataCenter, graphSage, classification, ds, device, args.max_vali_f1, args.name)
if args.learn_method != 'unsup':
args.max_vali_f1 = evaluate(dataCenter, ds, graphSage, classification, device, args.max_vali_f1, args.name, epoch)3 加载数据集部分 src/dataCenter.py
首先__init__()
读取数据load_dataSet()
import sys
import os
from collections import defaultdict
import numpy as np
class DataCenter(object):
"""docstring for DataCenter"""
def __init__(self, config):
super(DataCenter, self).__init__()
self.config = config
def load_dataSet(self, dataSet='cora'):
if dataSet == 'cora':
# cora_content_file = self.config['file_path.cora_content']
# cora_cite_file = self.config['file_path.cora_cite']
cora_content_file = '/Users/qiaoboyu/Desktop/pythonProject1/graphSAGE-pytorch-master/cora/cora.content'
cora_cite_file = '/Users/qiaoboyu/Desktop/pythonProject1/graphSAGE-pytorch-master/cora/cora.cites'
feat_data = []
labels = [] # label sequence of node
node_map = {} # map node to Node_ID
label_map = {} # map label to Label_ID
with open(cora_content_file) as fp:
for i,line in enumerate(fp):
info = line.strip().split()
feat_data.append([float(x) for x in info[1:-1]])
node_map[info[0]] = i
if not info[-1] in label_map:
label_map[info[-1]] = len(label_map)
labels.append(label_map[info[-1]])
# (2708, 1433)
feat_data = np.asarray(feat_data)
# (2708,)
labels = np.asarray(labels, dtype=np.int64)
adj_lists = defaultdict(set)
with open(cora_cite_file) as fp:
for i,line in enumerate(fp):
info = line.strip().split()
assert len(info) == 2
paper1 = node_map[info[0]]
paper2 = node_map[info[1]]
adj_lists[paper1].add(paper2) # defaultdict(set, {163: {402, 659}, 402: {163}})
adj_lists[paper2].add(paper1)
assert len(feat_data) == len(labels) == len(adj_lists) # 2708
test_indexs, val_indexs, train_indexs = self._split_data(feat_data.shape[0])
setattr(self, dataSet+'_test', test_indexs)
setattr(self, dataSet+'_val', val_indexs)
setattr(self, dataSet+'_train', train_indexs)
setattr(self, dataSet+'_feats', feat_data)
setattr(self, dataSet+'_labels', labels)
setattr(self, dataSet+'_adj_lists', adj_lists)
elif dataSet == 'pubmed':
pubmed_content_file = self.config['file_path.pubmed_paper']
pubmed_cite_file = self.config['file_path.pubmed_cites']
feat_data = []
labels = [] # label sequence of node
node_map = {} # map node to Node_ID
with open(pubmed_content_file) as fp:
fp.readline()
feat_map = {entry.split(":")[1]:i-1 for i,entry in enumerate(fp.readline().split("\t"))}
for i, line in enumerate(fp):
info = line.split("\t")
node_map[info[0]] = i
labels.append(int(info[1].split("=")[1])-1)
tmp_list = np.zeros(len(feat_map)-2)
for word_info in info[2:-1]:
word_info = word_info.split("=")
tmp_list[feat_map[word_info[0]]] = float(word_info[1])
feat_data.append(tmp_list)
feat_data = np.asarray(feat_data)
labels = np.asarray(labels, dtype=np.int64)
adj_lists = defaultdict(set)
with open(pubmed_cite_file) as fp:
fp.readline()
fp.readline()
for line in fp:
info = line.strip().split("\t")
paper1 = node_map[info[1].split(":")[1]]
paper2 = node_map[info[-1].split(":")[1]]
adj_lists[paper1].add(paper2)
adj_lists[paper2].add(paper1)
assert len(feat_data) == len(labels) == len(adj_lists)
test_indexs, val_indexs, train_indexs = self._split_data(feat_data.shape[0])
setattr(self, dataSet+'_test', test_indexs)
setattr(self, dataSet+'_val', val_indexs)
setattr(self, dataSet+'_train', train_indexs)
setattr(self, dataSet+'_feats', feat_data)
setattr(self, dataSet+'_labels', labels)
setattr(self, dataSet+'_adj_lists', adj_lists)分割数据集;
def _split_data(self, num_nodes, test_split = 3, val_split = 6):
rand_indices = np.random.permutation(num_nodes)
test_size = num_nodes // test_split #902
val_size = num_nodes // val_split # 451
train_size = num_nodes - (test_size + val_size) # 1355
test_indexs = rand_indices[:test_size] #随机打乱的序号
val_indexs = rand_indices[test_size:(test_size+val_size)]
train_indexs = rand_indices[(test_size+val_size):]
return test_indexs, val_indexs, train_indexs4 定义graphsage模型models.py
首先是定义__init__() 方法
class GraphSage(nn.Module):
"""docstring for GraphSage"""
def __init__(self, num_layers, input_size, out_size, raw_features, adj_lists, device, gcn=False, agg_func='MEAN'):
super(GraphSage, self).__init__()
self.input_size = input_size # 1433
self.out_size = out_size #128
self.num_layers = num_layers #2
self.gcn = gcn # False
self.device = device
self.agg_func = agg_func # MEAN 聚合函数(采样后要聚合,这里采用mean的方法)
self.raw_features = raw_features #torch.Size([2708, 1433]) 点的特征
self.adj_lists = adj_lists # 边的连接,现在还不是邻接矩阵
for index in range(1, num_layers+1): # graphsage每一层的构造
layer_size = out_size if index != 1 else input_size # 第一层应该是原始维度,第二层可以是更新后的维度
setattr(self, 'sage_layer'+str(index), SageLayer(layer_size, out_size, gcn=self.gcn)) # SageLayer类 定义图的每一层权重的定义,第一层 W = [128, 1433*2]
第二层 W = [128, 128*2]
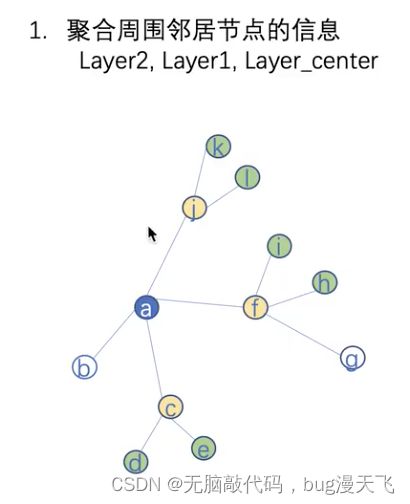
比如聚合节点a的表示,首先第一层聚合标绿色节点部分,得到128维向量,即黄色节点部分;
第二层聚合黄色节点部分,最终得到节点a的聚合后表示;
定义forward()
def forward(self, nodes_batch):
"""
Generates embeddings for a batch of nodes.
nodes_batch -- batch of nodes to learn the embeddings
"""
# 将节点转化为list类型
lower_layer_nodes = list(nodes_batch)
# 第一次放入的节点
nodes_batch_layers = [(lower_layer_nodes,)]
# self.dc.logger.info('get_unique_neighs.')
# 遍历每一层的graphsage
for i in range(self.num_layers):
# 得到初始节点的邻域节点_get_unique_neighs_list
lower_samp_neighs, lower_layer_nodes_dict, lower_layer_nodes= self._get_unique_neighs_list(lower_layer_nodes)
# 将得到的表示插入到nodes_batch_layers中
# 是采用向前插入的方式
# 首先要是中心节点,然后是与中心节点连接的节点部分,然后是与连接节点连接的部分
# 因此要把最外层的节点插入到最前面
# layer1, layer0,layer_center
nodes_batch_layers.insert(0, (lower_layer_nodes, lower_samp_neighs, lower_layer_nodes_dict))
# 层数加1,表示最开始的节点
assert len(nodes_batch_layers) == self.num_layers + 1
# 所有节点特征赋予到变量中
pre_hidden_embs = self.raw_features
for index in range(1, self.num_layers+1):
# 先选取nodes_batch_layers第0层对应的节点是哪些(layer0)
# nodes_batch_layers = [layer1,layer0,layer_center]
#
nb = nodes_batch_layers[index][0]
# 取nodes_batch_layers第1层对应的节点是哪些(layer1)
pre_neighs = nodes_batch_layers[index-1]
# self.dc.logger.info('aggregate_feats.')
# 聚合邻居节点和其中心节点
aggregate_feats = self.aggregate(nb, pre_hidden_embs, pre_neighs)
# 取对应的sage_layer
sage_layer = getattr(self, 'sage_layer'+str(index))
if index > 1:
# 第一层的batch节点没有进行转换,要进行一下转换
nb = self._nodes_map(nb, pre_hidden_embs, pre_neighs)
# self.dc.logger.info('sage_layer.')
# 开始图层之间的聚合操作 输入中心节点特征和聚合之后的特征
cur_hidden_embs = sage_layer(self_feats=pre_hidden_embs[nb],
aggregate_feats=aggregate_feats)
# 经过第一层graphsage后的表示为:[2157,128], 2157是节点数量
pre_hidden_embs = cur_hidden_embs
return pre_hidden_embs
def _nodes_map(self, nodes, hidden_embs, neighs):
#
layer_nodes, samp_neighs, layer_nodes_dict = neighs
assert len(samp_neighs) == len(nodes)
index = [layer_nodes_dict[x] for x in nodes] # 记录上一层的节点编码
return index
def _get_unique_neighs_list(self, nodes, num_sample=10): # num_sample 采样数
#nodes 1024个,导入每个batch的节点
_set = set
# adj_lists 是每一个点所连接的节点有哪些
# to_neighs 得到每个节点所属邻居的列表
to_neighs = [self.adj_lists[int(node)] for node in nodes] # len(nodes): 1024 节点邻居
# num_sample 周围采样邻居的数量,而不是对所有邻居都进行采样
if not num_sample is None: #首先对每一个节点的邻居集合neigh进行遍历,判断一下已有邻居数和采样数大小,多于采样数进行抽样
_sample = random.sample
# 遍历所有的to_neighs
# 如果to_neigh长度大于num_sample,则对其进行采样,如果小于num_sample 则放入所有节点
samp_neighs = [_set(_sample(to_neigh, num_sample)) if len(to_neigh) >= num_sample else to_neigh for to_neigh in to_neighs]
else:
samp_neighs = to_neighs
samp_neighs = [samp_neigh | set([nodes[i]]) for i, samp_neigh in enumerate(samp_neighs)] #聚合邻居节点信息时加上自己本身节点的信息
# 得到涉及到的节点的数量
_unique_nodes_list = list(set.union(*samp_neighs)) #把一个批次内的所有节点的邻居节点编号聚集在一块并去重
# 得到一个重新的排列
i = list(range(len(_unique_nodes_list)))
# 得到字典编号,节点对应的编号
unique_nodes = dict(list(zip(_unique_nodes_list, i))) #为所有的邻居节点建立一个索引映射
# samp_neighs 所有邻居节点的集合;
# unique_nodes 所有节点对应的字典
# _unique_nodes_list 所有节点的列表
return samp_neighs, unique_nodes, _unique_nodes_list
def aggregate(self, nodes, pre_hidden_embs, pre_neighs, num_sample=10):
# 取出最外层节点的信息,对应图中最外层的绿色节点
unique_nodes_list, samp_neighs, unique_nodes = pre_neighs
assert len(nodes) == len(samp_neighs)
# 节点本身是否包含着邻居节点中,也就是黄色节点+绿色节点
indicator = [(nodes[i] in samp_neighs[i]) for i in range(len(samp_neighs))]
assert (False not in indicator)
if not self.gcn:
# 去掉要进行聚合表示的节点(也就是中心节点本身,图中对应为去掉黄色节点)
samp_neighs = [(samp_neighs[i]-set([nodes[i]])) for i in range(len(samp_neighs))]
# self.dc.logger.info('2')
# 如果涉及到所有节点,则保留原矩阵;
# 如果不涉及所有节点,保留部分矩阵
if len(pre_hidden_embs) == len(unique_nodes):
embed_matrix = pre_hidden_embs
else:
embed_matrix = pre_hidden_embs[torch.LongTensor(unique_nodes_list)]
# self.dc.logger.info('3')
# 初始化了全为0的邻接矩阵(有关系的节点构成的邻接矩阵,不是所有节点)
# 本层节点数量,涉及到上层节点数量
mask = torch.zeros(len(samp_neighs), len(unique_nodes))
column_indices = [unique_nodes[n] for samp_neigh in samp_neighs for n in samp_neigh]
row_indices = [i for i in range(len(samp_neighs)) for j in range(len(samp_neighs[i]))]
# 有连接的点的值赋予成1
mask[row_indices, column_indices] = 1
# self.dc.logger.info('4')
if self.agg_func == 'MEAN':
# 按行求和,保持和输入一个维度
num_neigh = mask.sum(1, keepdim=True)
# 归一化操作,按行
mask = mask.div(num_neigh).to(embed_matrix.device)
# 矩阵相乘,相当于聚合周围邻居节点的信息求和
aggregate_feats = mask.mm(embed_matrix)
elif self.agg_func == 'MAX':
# print(mask)
indexs = [x.nonzero() for x in mask==1]
aggregate_feats = []
# self.dc.logger.info('5')
for feat in [embed_matrix[x.squeeze()] for x in indexs]:
if len(feat.size()) == 1:
aggregate_feats.append(feat.view(1, -1))
else:
aggregate_feats.append(torch.max(feat,0)[0].view(1, -1))
aggregate_feats = torch.cat(aggregate_feats, 0)
# self.dc.logger.info('6')
# 返回聚合之后的特征
return aggregate_feats5 定义SageLayer src/models.py
定义__init__()
class SageLayer(nn.Module):
"""
Encodes a node's using 'convolutional' GraphSage approach
"""
def __init__(self, input_size, out_size, gcn=False):
super(SageLayer, self).__init__()
self.input_size = input_size # 1433
self.out_size = out_size # 128
self.gcn = gcn
# 初始化,连接操作,设置为2倍 torch.Size([128, 2866])
# 定义要学习的参数
self.weight = nn.Parameter(torch.FloatTensor(out_size, self.input_size if self.gcn else 2 * self.input_size))
# 这里设置成2*input_size是因为这一层的表示将上一层用户节点的嵌入与其邻居节点的嵌入连接到了一起,如下图所示
self.init_params() # 初始化参数
def init_params(self):
for param in self.parameters():
nn.init.xavier_uniform_(param)6 定义分类器模型 src/models.py
定义初始化函数__init__()
class Classification(nn.Module):
def __init__(self, emb_size, num_classes):
super(Classification, self).__init__()
#self.weight = nn.Parameter(torch.FloatTensor(emb_size, num_classes))
self.layer = nn.Sequential(
nn.Linear(emb_size, num_classes)
#nn.ReLU()
)
self.init_params()
def init_params(self):
for param in self.parameters():
if len(param.size()) == 2:
nn.init.xavier_uniform_(param)定义forward();
def forward(self, self_feats, aggregate_feats, neighs=None):
"""
Generates embeddings for a batch of nodes.
nodes -- list of nodes
"""
if not self.gcn:
# 节点特征及节点周围邻居特征连接在一起
combined = torch.cat([self_feats, aggregate_feats], dim=1)
else:
combined = aggregate_feats
# 与可学习权重相乘
combined = F.relu(self.weight.mm(combined.t())).t()
return combined7 模型运行函数 src/utils.py
def apply_model(dataCenter, ds, graphSage, classification, unsupervised_loss, b_sz, unsup_loss, device, learn_method):
# 验证集、测试集、训练集节点特征及label
test_nodes = getattr(dataCenter, ds+'_test')
val_nodes = getattr(dataCenter, ds+'_val')
train_nodes = getattr(dataCenter, ds+'_train')
labels = getattr(dataCenter, ds+'_labels')
# 无监督的loss, 这里采用有监督,因此该定义无影响
if unsup_loss == 'margin':
num_neg = 6
elif unsup_loss == 'normal':
num_neg = 100
else:
print("unsup_loss can be only 'margin' or 'normal'.")
sys.exit(1)
# 打乱训练集
train_nodes = shuffle(train_nodes)
# 定义模型
models = [graphSage, classification]
# 定义模型参数
params = []
# 循环模型
for model in models:
# 遍历模型所有参数
for param in model.parameters():
# 参数定义为可训练的梯度
if param.requires_grad:
params.append(param) # W和bias
optimizer = torch.optim.SGD(params, lr=0.7)
optimizer.zero_grad()
# 初始化模型梯度
for model in models:
model.zero_grad()
# 每一轮训练迭代数
# b_sz定义为20
batches = math.ceil(len(train_nodes) / b_sz)
visited_nodes = set()
# 遍历每一个batch
for index in range(batches):
# batch内的节点
nodes_batch = train_nodes[index*b_sz:(index+1)*b_sz]
# extend nodes batch for unspervised learning
# no conflicts with supervised learning
# 对于无监督,在无监督上进行了负采样的操作
# 对于有监督命令的执行是不冲突的 ,只是训练节点的数量增加了, 这里是1024个节点
nodes_batch = np.asarray(list(unsupervised_loss.extend_nodes(nodes_batch, num_neg=num_neg)))
visited_nodes |= set(nodes_batch)
# get ground-truth for the nodes batch
# 得到节点的label
labels_batch = labels[nodes_batch]
# feed nodes batch to the graphSAGE
# returning the nodes embeddings
# 跳入到graphsage层中,学习到节点表征
# [1024,128]
embs_batch = graphSage(nodes_batch)
if learn_method == 'sup':
# superivsed learning
# 得到[1024,7]
logists = classification(embs_batch)
loss_sup = -torch.sum(logists[range(logists.size(0)), labels_batch], 0)
loss_sup /= len(nodes_batch)
loss = loss_sup
elif learn_method == 'plus_unsup':
# superivsed learning
logists = classification(embs_batch)
loss_sup = -torch.sum(logists[range(logists.size(0)), labels_batch], 0)
loss_sup /= len(nodes_batch)
# unsuperivsed learning
if unsup_loss == 'margin':
loss_net = unsupervised_loss.get_loss_margin(embs_batch, nodes_batch)
elif unsup_loss == 'normal':
loss_net = unsupervised_loss.get_loss_sage(embs_batch, nodes_batch)
loss = loss_sup + loss_net
else:
if unsup_loss == 'margin':
loss_net = unsupervised_loss.get_loss_margin(embs_batch, nodes_batch)
elif unsup_loss == 'normal':
loss_net = unsupervised_loss.get_loss_sage(embs_batch, nodes_batch)
loss = loss_net
print('Step [{}/{}], Loss: {:.4f}, Dealed Nodes [{}/{}] '.format(index+1, batches, loss.item(), len(visited_nodes), len(train_nodes)))
loss.backward()
for model in models:
nn.utils.clip_grad_norm_(model.parameters(), 5)
optimizer.step()
optimizer.zero_grad()
for model in models:
model.zero_grad()
return graphSage, classification想要更详细,看B站的视频,源码+调试的讲,很清楚!!
16. 4.3_GraphSAGE代码_哔哩哔哩_bilibili