mlr3绘制校准曲线
本文首发于公众号:医学和生信笔记
“医学和生信笔记,专注R语言在临床医学中的使用,R语言数据分析和可视化。主要分享R语言做医学统计学、meta分析、网络药理学、临床预测模型、机器学习、生物信息学等。
前面介绍了使用tidymodels画校准曲线,不知道大家学会了没?
众所周知,tidymodels目前还不支持一键绘制校准曲线!相同类型的mlr3也是不支持的!大家多去github提issue,加速对校准曲线的支持!
今天介绍mlr3怎么画校准曲线,还是那句话,校准曲线就是散点图,你非说是折线图也行......
加载R包
首先还是加载数据和R包,和之前的数据一样的。
library(mlr3verse)
## Loading required package: mlr3
library(mlr3pipelines)
library(mlr3filters)
建立任务
然后是对数据进行划分训练集和测试集,对数据进行预处理,为了和之前的tidymodels进行比较,这里使用的数据和预处理步骤都是和之前一样的。
# 读取数据
all_plays <- readRDS("../000files/all_plays.rds")
# 建立任务
pbp_task <- as_task_classif(all_plays, target="play_type")
# 数据划分
split_task <- partition(pbp_task, ratio=0.75)
task_train <- pbp_task$clone()$filter(split_task$train)
task_test <- pbp_task$clone()$filter(split_task$test)
数据预处理
建立任务后就是建立数据预处理步骤,这里采用和上篇推文tidymodels中一样的预处理步骤:
# 数据预处理
pbp_prep <- po("select", # 去掉3列
selector = selector_invert(
selector_name(c("half_seconds_remaining","yards_gained","game_id")))
) %>>%
po("colapply", # 把这两列变成因子类型
affect_columns = selector_name(c("posteam","defteam")),
applicator = as.factor) %>>%
po("filter", # 去除高度相关的列
filter = mlr3filters::flt("find_correlation"), filter.cutoff=0.3) %>>%
po("scale", scale = F) %>>% # 中心化
po("removeconstants") # 去掉零方差变量
建立模型
先选择随机森林模型。
rf_glr <- as_learner(pbp_prep %>>% lrn("classif.ranger", predict_type="prob"))
rf_glr$id <- "randomForest"
很多人喜欢在训练集中使用10折交叉验证,但其实这对于提高模型表现没什么用~尤其是临床预测模型这个领域~因为你的模型表现好不好很大程度上取决于你的数据好不好!鸭子是不会变成天鹅的
rr <- resample(task = task_train,
learner = rf_glr,
resampling = rsmp("cv",folds = 10),
store_models = T)
## INFO [18:25:28.412] [mlr3] Applying learner 'randomForest' on task 'all_plays' (iter 1/10)
## INFO [18:25:58.497] [mlr3] Applying learner 'randomForest' on task 'all_plays' (iter 2/10)
## INFO [18:26:29.302] [mlr3] Applying learner 'randomForest' on task 'all_plays' (iter 3/10)
## INFO [18:27:02.512] [mlr3] Applying learner 'randomForest' on task 'all_plays' (iter 4/10)
## INFO [18:27:31.100] [mlr3] Applying learner 'randomForest' on task 'all_plays' (iter 5/10)
## INFO [18:28:01.090] [mlr3] Applying learner 'randomForest' on task 'all_plays' (iter 6/10)
## INFO [18:28:30.868] [mlr3] Applying learner 'randomForest' on task 'all_plays' (iter 7/10)
## INFO [18:29:01.464] [mlr3] Applying learner 'randomForest' on task 'all_plays' (iter 8/10)
## INFO [18:29:32.870] [mlr3] Applying learner 'randomForest' on task 'all_plays' (iter 9/10)
## INFO [18:30:03.747] [mlr3] Applying learner 'randomForest' on task 'all_plays' (iter 10/10)
评价模型
先看看在训练集中的表现。
混淆矩阵:
rr$prediction()$confusion
## truth
## response pass run
## pass 31932 9886
## run 8819 18345
混淆矩阵可视化:
autoplot(rr$prediction())
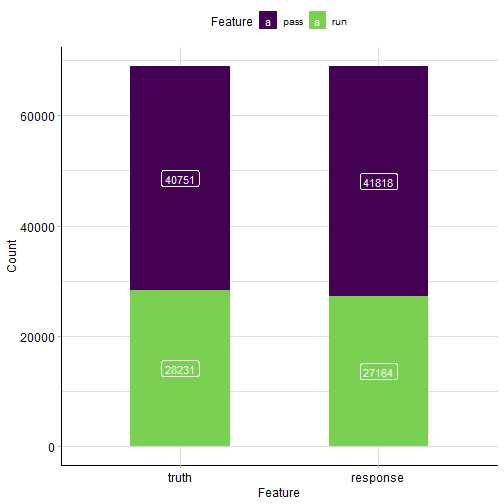 plot of chunk unnamed-chunk-7
plot of chunk unnamed-chunk-7
查看其他结果:
rr$aggregate(msrs(c("classif.auc","classif.acc","classif.bbrier")))
## classif.auc classif.acc classif.bbrier
## 0.7979179 0.7288424 0.1790592
喜闻乐见ROC曲线:
autoplot(rr,type = "roc")
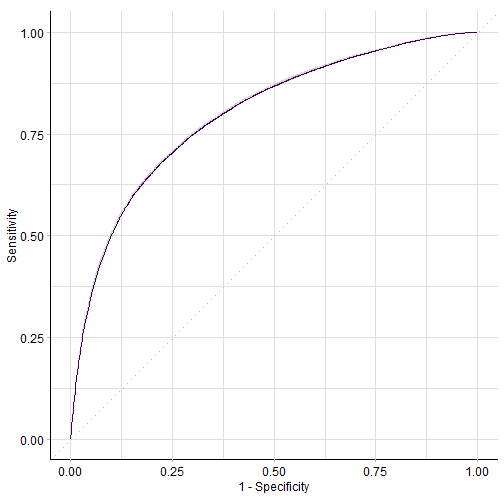 plot of chunk unnamed-chunk-9
plot of chunk unnamed-chunk-9
喜闻乐见的prc曲线:
autoplot(rr, type = "prc")
 plot of chunk unnamed-chunk-10
plot of chunk unnamed-chunk-10
箱线图:
autoplot(rr, measure = msr("classif.auc"))
 plot of chunk unnamed-chunk-11
plot of chunk unnamed-chunk-11
以上所有介绍的图形和评价方法都在之前的推文详细介绍过了~不会的赶紧翻看:mlr3实现多个模型评价和比较
训练集的校准曲线
先画训练集的校准曲线,毫无难度,看不懂的可以加群一起讨论~
prediction <- as.data.table(rr$prediction())
head(prediction)
## row_ids truth response prob.pass prob.run
## 1: 6 run run 0.4294702 0.57052982
## 2: 30 pass pass 0.7730236 0.22697638
## 3: 48 run run 0.2052662 0.79473378
## 4: 94 pass pass 0.6593303 0.34066970
## 5: 106 pass pass 0.5731238 0.42687625
## 6: 108 pass pass 0.9365055 0.06349447
方法1:
suppressPackageStartupMessages(library(tidyverse))
library(ggsci)
calibration_df <- prediction %>%
mutate(pass = if_else(truth == "pass", 1, 0),
pred_rnd = round(prob.pass, 2)
) %>%
group_by(pred_rnd) %>%
summarize(mean_pred = mean(prob.pass),
mean_obs = mean(pass),
n = n()
)
ggplot(calibration_df, aes(mean_pred, mean_obs))+
geom_point(aes(size = n), alpha = 0.5)+
scale_color_lancet()+
geom_abline(linetype = "dashed")+
labs(x="Predicted Probability", y= "Observed Probability")+
theme_minimal()
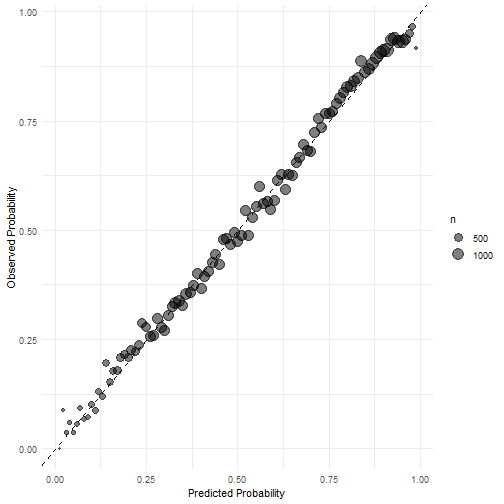 plot of chunk unnamed-chunk-13
plot of chunk unnamed-chunk-13
第2种方法,大家比较喜欢的折线图!
cali_df <- prediction %>%
arrange(prob.pass) %>%
mutate(pass = if_else(truth == "pass", 1, 0),
group = c(rep(1:100,each=680), rep(101,982))
) %>%
group_by(group) %>%
summarise(mean_pred = mean(prob.pass),
mean_obs = mean(pass)
)
ggplot(cali_df, aes(mean_pred, mean_obs))+
geom_line(size=1)+
labs(x="Predicted Probability", y= "Observed Probability")+
theme_minimal()
 plot of chunk unnamed-chunk-14
plot of chunk unnamed-chunk-14
是不是和上一篇中的tidymodels画出来的一模一样?没错,就是一样的,就是这么简单,想怎么画就怎么画 !
训练集的校准曲线
先把模型用在测试集上,得到预测结果,然后画图!
cv_pred <- rf_glr$train(task_train)$predict(task_test)
cv_pred_df <- as.data.table(cv_pred)
head(cv_pred_df)
row_ids truth response prob.pass prob.run
1: 2 pass run 0.4213731 0.5786269
2: 5 pass pass 0.8475027 0.1524973
3: 6 run run 0.3782730 0.6217270
4: 12 pass pass 0.6308144 0.3691856
5: 14 pass pass 0.8371294 0.1628706
6: 15 run run 0.1837391 0.8162609
先画个喜闻乐见的校准曲线:
cali_df <- cv_pred_df %>%
arrange(prob.pass) %>%
mutate(pass = if_else(truth == "pass", 1, 0),
group = c(rep(1:100,each=229), rep(101,94))
) %>%
group_by(group) %>%
summarise(mean_pred = mean(prob.pass),
mean_obs = mean(pass)
)
ggplot(cali_df, aes(mean_pred, mean_obs))+
geom_line(size=1)+
labs(x="Predicted Probability", y= "Observed Probability")+
theme_minimal()
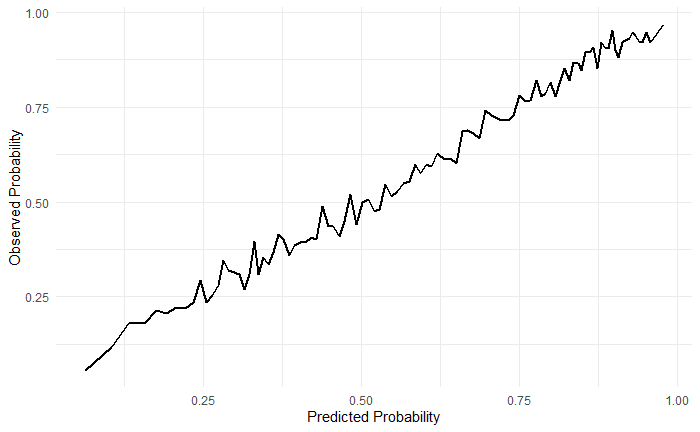
另一种颜值高点的校准曲线,给你点颜色瞧瞧!
calibration_df <- cv_pred_df %>%
mutate(pass = if_else(truth == "pass", 1, 0),
pred_rnd = round(prob.pass, 2)
) %>%
group_by(pred_rnd) %>%
summarize(mean_pred = mean(prob.pass),
mean_obs = mean(pass),
n = n()
) %>%
mutate(group = case_when(n < 100 ~ "<100",
n < 200 ~ "<200",
n < 300 ~ "<300",
n < 400 ~ "<400",
TRUE ~ "≥400"
))
## Error in mutate(., pass = if_else(truth == "pass", 1, 0), pred_rnd = round(prob.pass, : object 'cv_pred_df' not found
ggplot(calibration_df, aes(mean_pred, mean_obs))+
geom_point(aes(size = n, color = group))+
scale_color_jama()+
geom_abline(linetype = "dashed")+
labs(x="Predicted Probability", y= "Observed Probability")+
theme_minimal()
## Error in FUN(X[[i]], ...): object 'group' not found

配色略诡异...
“校准曲线,你学会了吗?
我知道并没有,比如,多条画一起怎么搞?生存资料的怎么搞?
关于这两个问题,可以翻看我之前的推文
加群即可免费获得今日示例数据!
本文首发于公众号:医学和生信笔记
“医学和生信笔记,专注R语言在临床医学中的使用,R语言数据分析和可视化。主要分享R语言做医学统计学、meta分析、网络药理学、临床预测模型、机器学习、生物信息学等。
本文由 mdnice 多平台发布