从sklearn.preprocessing, sklearn.feature_selection学习特征工程之预处理
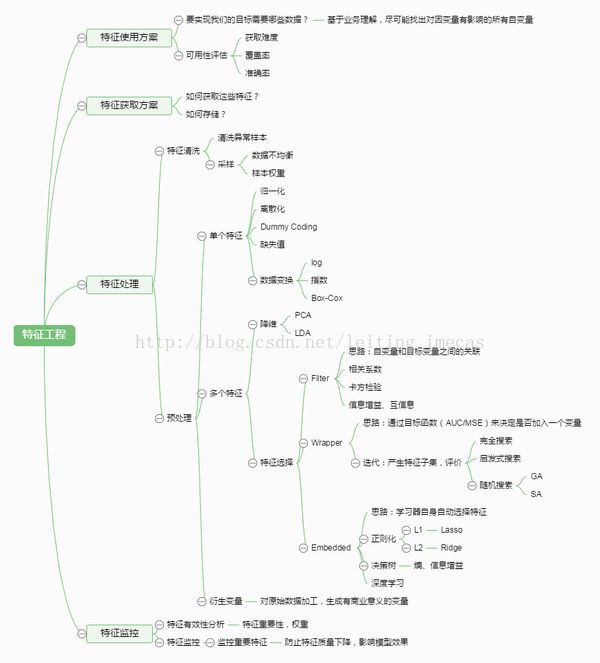
特征工程思维导图如下图。 本文借助sklearn介绍其中的预处理部分
二 单特征预处理
<1> 标准化 Standardization 或者叫 mean removal and variance scaling(平均值移除、方差缩放)
说明1: 标准化其实就是干两件事:“transform the data to center” ,即使数据平均值为0;
“scale it by dividing non-constant features by their standard deviation”,即标准差为1
(1) sklearn 提供了scale做数据标准化:
>>> from sklearn import preprocessing
>>> import numpy as np
>>> X = np.array([[1., -1., 2.],
... [2., 0., 0.],
... [0., 1., -1.]])
>>> X_scaled = preprocessing.scale(X)
>>> X_scaled
array([[ 0. , -1.22474487, 1.33630621],
[ 1.22474487, 0. , -0.26726124],
[-1.22474487, 1.22474487, -1.06904497]])
>>> X_scaled.mean(axis=0)
array([ 0., 0., 0.])
>>> X_scaled.std(axis=0)
array([ 1., 1., 1.])preprocessing 提供了另一个实用的类StandardScaler,它的fit函数解析训练数据(包括值和矩阵格式),transform则对
数据执行standardization过程。 在训练数据上使用的StandardScalar对象可以再使用在测试数据上
>>> scaler = preprocessing.StandardScaler().fit(X)
>>> scaler
StandardScaler(copy=True, with_mean=True, with_std=True)
>>> scaler.mean_
array([ 1. , 0. , 0.33333333])
>>> scaler.scale_
array([ 0.81649658, 0.81649658, 1.24721913])
>>> scaler.transform(X)
array([[ 0. , -1.22474487, 1.33630621],
[ 1.22474487, 0. , -0.26726124],
[-1.22474487, 1.22474487, -1.06904497]])
>>> X_test = np.array([[-1., 1., 0.]])
>>> scaler.transform(X_test)
array([[-2.44948974, 1.22474487, -0.26726124]])(2) 缩放至一定范围
先介绍MinMaxScaler的使用, 使用max - min作为分母
>>> X_train = np.array([[1., -1., 2.],
... [2., 0., 0.],
... [0., 1., -1]])
>>> min_max_scaler = preprocessing.MinMaxScaler()
>>> X_train_minmax = min_max_scaler.fit_transform(X_train)
>>> X_train_minmax
array([[ 0.5 , 0. , 1. ],
[ 1. , 0.5 , 0.33333333],
[ 0. , 1. , 0. ]])
>>> X_test = np.array([[ -3., -1., 4.]])
>>> X_test_minmax = min_max_scaler.transform(X_test)
>>> X_test_minmax
array([[-1.5 , 0. , 1.66666667]])在介绍MaxAbsScaler, 将最大的绝对值作为分母
>>> max_abs_scaler = preprocessing.MaxAbsScaler()
>>> X_train_maxabs = max_abs_scaler.fit_transform(X_train)
>>> X_train_maxabs
array([[ 0.5, -1. , 1. ],
[ 1. , 0. , 0. ],
[ 0. , 1. , -0.5]])
>>> X_test = np.array([[ -3., -1., 4.]])
>>> X_test_maxabs = max_abs_scaler.transform(X_test)
>>> X_test_maxabs
array([[-1.5, -1. , 2. ]])
>>> max_abs_scaler.scale_
array([ 2., 1., 2.])<2> 正则化 Normalization (跟以前理解的正则化概念不同)
正则化的过程是将每个样本缩放到单位范数。对于后面要通过点积或其他核方法计算两个样本之间相似性的情况,
该正则化十分有用。
Normalization主要思想是对每个样本计算p-范数(例如l1-范数,l2-范数等),然后样本的每个元素除以该范数。则
执行正则化的样本的p-范数变为1。 p-范数的公式: ||X||p = (|x1|^p + |x2|^p + .... + |xn|^p)^1/p
该方法主要用于文本分类和聚类,例如两个tf-idf向量的l2正则化进行点积就得到两个向量的余弦相似性
>>> X_normalized = preprocessing.normalize(X, norm='l2')>>> X_normalized
array([[ 0.40824829, -0.40824829, 0.81649658],
[ 1. , 0. , 0. ],
[ 0. , 0.70710678, -0.70710678]])同样提供了类:
>>> normalizer = preprocessing.Normalizer().fit(X)
>>> normalizer
Normalizer(copy=True, norm='l2')
>>> normalizer.transform(X)
array([[ 0.40824829, -0.40824829, 0.81649658],
[ 1. , 0. , 0. ],
[ 0. , 0.70710678, -0.70710678]])
>>> normalizer.transform([[-1., 1., 0.]])
array([[-0.70710678, 0.70710678, 0. ]])<3> Binarization 二值化
特征二值化是指依据一个阈值,将大于阈值的赋值为1, 小于等于阈值的赋值为0.
>>> X
array([[ 1., -1., 2.],
[ 2., 0., 0.],
[ 0., 1., -1.]])
>>> binarizer = preprocessing.Binarizer().fit(X)
>>> binarizer
Binarizer(copy=True, threshold=0.0)
>>> binarizer.transform(X)
array([[ 1., 0., 1.],
[ 1., 0., 0.],
[ 0., 1., 0.]])
>>> binarizer2 = preprocessing.Binarizer(threshold=1.1)
>>> binarizer2.transform(X)
array([[ 0., 0., 1.],
[ 1., 0., 0.],
[ 0., 0., 0.]])<4> 类别型特征编码
可以使用oneHot encoding 将类别型特征进行编码。
>>> enc = preprocessing.OneHotEncoder()
>>> enc.fit([[0, 0, 3], [1, 1, 0], [0, 2, 1], [1, 0, 2]]) # 第一个特征有2个类别,第二个特征有3个类别,第三个特征有4类别
OneHotEncoder(categorical_features='all', dtype=,
handle_unknown='error', n_values='auto', sparse=True)
>>> enc.transform([[0, 0, 0]]).toarray()
array([[ 1., 0., 1., 0., 0., 1., 0., 0., 0.]]) # 第一个特征占据前两位,第二个特征占据厚3个位置,第三个特征占最后4个
>>> enc.transform([[1, 0, 0]]).toarray()
array([[ 0., 1., 1., 0., 0., 1., 0., 0., 0.]])
>>> enc.transform([[0, 1, 0]]).toarray()
array([[ 1., 0., 0., 1., 0., 1., 0., 0., 0.]])
>>> enc.transform([[0, 2, 0]]).toarray()
array([[ 1., 0., 0., 0., 1., 1., 0., 0., 0.]])
>>> enc.transform([[0, 0, 1]]).toarray()
array([[ 1., 0., 1., 0., 0., 0., 1., 0., 0.]])
>>> enc.transform([[0, 0, 2]]).toarray()
array([[ 1., 0., 1., 0., 0., 0., 0., 1., 0.]])
>>> enc.transform([[0, 0, 3]]).toarray()
array([[ 1., 0., 1., 0., 0., 0., 0., 0., 1.]]) 如果训练数据中的类别不全,则可以指定每个特征的类别数量
>>> enc = preprocessing.OneHotEncoder(n_values=[2,3,4]) #指定
>>> enc.fit([[1, 2, 3], [0, 2, 0]])
OneHotEncoder(categorical_features='all', dtype=,
handle_unknown='error', n_values=[2, 3, 4], sparse=True)
>>> enc.transform([[1, 2, 0]]).toarray()
array([[ 0., 1., 0., 0., 1., 1., 0., 0., 0.]])
>>> enc.transform([[1, 0, 0]]).toarray()
array([[ 0., 1., 1., 0., 0., 1., 0., 0., 0.]])
>>> enc.transform([[1, 1, 0]]).toarray()
array([[ 0., 1., 0., 1., 0., 1., 0., 0., 0.]]) <5> 缺省值计算 ---------Imputer
>>> from sklearn.preprocessing import Imputer
>>> imp = Imputer(missing_values='NaN', strategy='mean', axis=0)
>>> imp
Imputer(add_indicator_features=False, axis=0, copy=True, missing_values='NaN',
strategy='mean', verbose=0)
>>> imp.fit([[1, 2], [np.nan, 3], [7, 6]]) #第一维的平均值为4, 第二维的平均值为3.667
Imputer(add_indicator_features=False, axis=0, copy=True, missing_values='NaN',
strategy='mean', verbose=0)
>>> X = [[np.nan, 2], [6, np.nan], [7, 6]]
>>> imp.transform(X)
array([[ 4. , 2. ],
[ 6. , 3.66666667],
[ 7. , 6. ]])<6> 特征的多项式变换
多项式变换会将每个样本数据扩展为原本元素的多项式项,例如
(X1, X2) 经过二项式变换,变为(1, X1, X2, X1^2, X1X2, X2^2)
>>> X = np.arange(6).reshape(3,2)
>>> X
array([[0, 1],
[2, 3],
[4, 5]])
>>> poly = preprocessing.PolynomialFeatures(2) #指定二项式变化
>>> poly.fit_transform(X)
array([[ 1., 0., 1., 0., 0., 1.],
[ 1., 2., 3., 4., 6., 9.],
[ 1., 4., 5., 16., 20., 25.]])还可以指定只保留互动项:
(X1, X2) 经过二项式变换只保留互动项,变为(1, X1, X2, X1X2)
>>> X
array([[0, 1],
[2, 3],
[4, 5]])
>>> poly2 = preprocessing.PolynomialFeatures(degree=2, interaction_only=True)
>>> poly2.fit_transform(X)
array([[ 1., 0., 1., 0.],
[ 1., 2., 3., 6.],
[ 1., 4., 5., 20.]])<7> 自定义变换
可是使用FunctionTransformer指定一个子选择的变换方法
>>> from numpy import log1p # log1p(x) = log(1 + x)
>>> from sklearn.preprocessing import FunctionTransformer
>>> trans = FunctionTransformer(log1p)
>>> X = np.array([[0,1], [2, 3]])
>>> trans.transform(X)
array([[ 0. , 0.69314718],
[ 1.09861229, 1.38629436]])三 特征选择
<1> Filter 过滤法 ---------多使用于线性的模型
1.1 方差选择法
>>> from sklearn.feature_selection import VarianceThreshold
>>> X = [[0, 0, 1], [0, 1, 0], [1, 0, 0], [0, 1, 1], [0, 1, 0], [0, 1, 1]]
>>> sel = VarianceThreshold(threshold=(.8 * (1 - .8))) #
>>> sel.fit_transform(X)
array([[0, 1],
[1, 0],
[0, 0],
[1, 1],
[1, 0],
[1, 1]])1.2 Univariate feature selection------ 单变量特征选择
1.2.1 SelectKBest ------保留k个最好的特征。 打分方法,例如卡方验证
1.2.2 SelectPercentile------- 保留一定比例
>>> from sklearn.datasets import load_iris
>>> from sklearn.feature_selection import SelectKBest
>>> from sklearn.feature_selection import chi2
>>> iris = load_iris()
>>> X, y = iris.data, iris.target
>>> X.shape
(150, 4)
>>> X_new = SelectKBest(chi2, k=2).fit_transform(X, y)
>>> X_new.shape
(150, 2)<2> Wrapper --- 包裹法
sklearn.feature_selection.RFE(estimator, n_features_to_select=None, step=1, verbose=0)
>>> from sklearn.feature_selection import RFE
>>> from sklearn.linear_model import LogisticRegression
>>> rfe = RFE(LogisticRegression(), n_features_to_select=1, step=1)
>>> rfe.fit(X, y)
<3> Embedded -----嵌入法, SelectFromModel
sklearn.feature_selection.SelectFromModel(estimator, threshold=None, prefit=False)(1) 基于L1正则化的特征选择方法
>>> from sklearn.svm import LinearSVC
>>> from sklearn.datasets import load_iris
>>> from sklearn.feature_selection import SelectFromModel
>>> iris = load_iris()
>>> X, y = iris.data, iris.target
>>> X.shape
(150, 4)
>>> lsvc = LinearSVC(C=0.01, penalty="l1", dual=False).fit(X,y)
>>> model = SelectFromModel(lsvc, prefit=True)
>>> X_new = model.transform(X)
>>> X_new.shape
(150, 3)>>> from sklearn.ensemble import ExtraTreesClassifier
>>> from sklearn.datasets import load_iris
>>> from sklearn.feature_selection import SelectFromModel
>>> iris = load_iris()
>>> X, y = iris.data, iris.target
>>> X.shape
(150, 4)
>>> clf = ExtraTreesClassifier()
>>> clf = clf.fit(X,y)
>>> clf.feature_importances_
array([ 0.05517225, 0.06978394, 0.52250383, 0.35253998]) #特征重要性
>>> model = SelectFromModel(clf, prefit=True)
>>> X_new = model.transform(X)
>>> X_new.shape
(150, 2)[source]