pytorch搭建MobileViT网络——一种用于移动设备的轻量级通用视觉 transformer
MobileViT介绍
论文地址:https://arxiv.org/pdf/2110.02178.pdf
来自苹果的研究者提出了一种用于移动设备的轻量级通用视觉 transformer——MobileViT。该网络在 ImageNet-1k 数据集上实现了 78.4% 的最佳精度,比 MobileNetv3 还要高 3.2%,而且训练方法简单。
轻量级卷积神经网络(CNN)是移动设备机器视觉任务的首选。它们的空间归纳偏置允许它们在不同的视觉任务中,可以使用较少的参数进行视觉内容的学习表示。但是这些网络在空间上是局部的。为了学习视觉内容的全局表示,要采用基于自注意力的视觉 transformer(ViT)。与 CNN 不同,ViT 是重量级的。在这篇文章中,作者提出了以下问题:是否有可能结合 CNN 和 ViT 的优势,为移动设备机器视觉任务构建一个轻量级、低延迟的神经网络模型?
为了解决上面的问题,作者提出了 MobileViT——一种用于移动设备的轻量级通用视觉 transformer。MobileViT 从另一个角度介绍了使用 transformer 进行全局信息处理的方法。
MobileViT结构
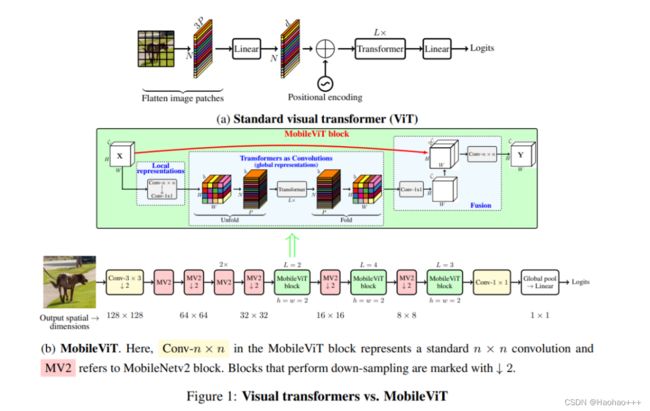
如图 b 所示的 MobileViT 块的作用是使用包含较少参数的输入张量学习局部和全局信息。MobileViT 应用一个 n×n 标准卷积层,然后是逐点(1×1)卷积层来特征提取。n×n 卷积层编码局部空间信息,而逐点卷积通过学习输入通道的线性组合将张量投影到高维空间。
class MobileViTBlock(nn.Module):
def __init__(self, dim, depth, channel, kernel_size, patch_size, mlp_dim, dropout=0.):
super().__init__()
self.ph, self.pw = patch_size
self.conv1 = conv_nxn_bn(channel, channel, kernel_size)
self.conv2 = conv_1x1_bn(channel, dim)
self.transformer = Transformer(dim, depth, 4, 8, mlp_dim, dropout)
self.conv3 = conv_1x1_bn(dim, channel)
self.conv4 = conv_nxn_bn(2 * channel, channel, kernel_size)
def forward(self, x):
y = x.clone()
# Local representations
x = self.conv1(x)
x = self.conv2(x)
# Global representations
_, _, h, w = x.shape
x = rearrange(x, 'b d (h ph) (w pw) -> b (ph pw) (h w) d', ph=self.ph, pw=self.pw)
x = self.transformer(x)
x = rearrange(x, 'b (ph pw) (h w) d -> b d (h ph) (w pw)', h=h//self.ph, w=w//self.pw, ph=self.ph, pw=self.pw)
# Fusion
x = self.conv3(x)
x = torch.cat((x, y), 1)
x = self.conv4(x)
return x
总而言之, MobileViT 使用标准卷积和 transformer 分别学习局部和全局表示,使得MobileViT 既具有类似卷积的属性,又同时允许全局处理。
代码实现
需要安装einops, einops库可以帮助我们很轻松的对张量进行调整,功能类似与我们平时使用的reshape、view、transpose和permute等操作,相信接触过深度学习的同学一定对这些函数比较的熟悉。
注意,输入图片不能是224x224,因为reshape时会报错,可以设置成256x256或其他。
import torch
import torch.nn as nn
from einops import rearrange
def conv_1x1_bn(inp, oup):
return nn.Sequential(
nn.Conv2d(inp, oup, 1, 1, 0, bias=False),
nn.BatchNorm2d(oup),
nn.SiLU()
)
def conv_nxn_bn(inp, oup, kernal_size=3, stride=1):
return nn.Sequential(
nn.Conv2d(inp, oup, kernal_size, stride, 1, bias=False),
nn.BatchNorm2d(oup),
nn.SiLU()
)
class PreNorm(nn.Module):
def __init__(self, dim, fn):
super().__init__()
self.norm = nn.LayerNorm(dim)
self.fn = fn
def forward(self, x, **kwargs):
return self.fn(self.norm(x), **kwargs)
class FeedForward(nn.Module):
def __init__(self, dim, hidden_dim, dropout=0.):
super().__init__()
self.net = nn.Sequential(
nn.Linear(dim, hidden_dim),
nn.SiLU(),
nn.Dropout(dropout),
nn.Linear(hidden_dim, dim),
nn.Dropout(dropout)
)
def forward(self, x):
return self.net(x)
class Attention(nn.Module):
def __init__(self, dim, heads=8, dim_head=64, dropout=0.):
super().__init__()
inner_dim = dim_head * heads
project_out = not (heads == 1 and dim_head == dim)
self.heads = heads
self.scale = dim_head ** -0.5
self.attend = nn.Softmax(dim = -1)
self.to_qkv = nn.Linear(dim, inner_dim * 3, bias = False)
self.to_out = nn.Sequential(
nn.Linear(inner_dim, dim),
nn.Dropout(dropout)
) if project_out else nn.Identity()
def forward(self, x):
qkv = self.to_qkv(x).chunk(3, dim=-1)
q, k, v = map(lambda t: rearrange(t, 'b p n (h d) -> b p h n d', h = self.heads), qkv)
dots = torch.matmul(q, k.transpose(-1, -2)) * self.scale
attn = self.attend(dots)
out = torch.matmul(attn, v)
out = rearrange(out, 'b p h n d -> b p n (h d)')
return self.to_out(out)
class Transformer(nn.Module):
def __init__(self, dim, depth, heads, dim_head, mlp_dim, dropout=0.):
super().__init__()
self.layers = nn.ModuleList([])
for _ in range(depth):
self.layers.append(nn.ModuleList([
PreNorm(dim, Attention(dim, heads, dim_head, dropout)),
PreNorm(dim, FeedForward(dim, mlp_dim, dropout))
]))
def forward(self, x):
for attn, ff in self.layers:
x = attn(x) + x
x = ff(x) + x
return x
class MV2Block(nn.Module):
def __init__(self, inp, oup, stride=1, expansion=4):
super().__init__()
self.stride = stride
assert stride in [1, 2]
hidden_dim = int(inp * expansion)
self.use_res_connect = self.stride == 1 and inp == oup
if expansion == 1:
self.conv = nn.Sequential(
# dw
nn.Conv2d(hidden_dim, hidden_dim, 3, stride, 1, groups=hidden_dim, bias=False),
nn.BatchNorm2d(hidden_dim),
nn.SiLU(),
# pw-linear
nn.Conv2d(hidden_dim, oup, 1, 1, 0, bias=False),
nn.BatchNorm2d(oup),
)
else:
self.conv = nn.Sequential(
# pw
nn.Conv2d(inp, hidden_dim, 1, 1, 0, bias=False),
nn.BatchNorm2d(hidden_dim),
nn.SiLU(),
# dw
nn.Conv2d(hidden_dim, hidden_dim, 3, stride, 1, groups=hidden_dim, bias=False),
nn.BatchNorm2d(hidden_dim),
nn.SiLU(),
# pw-linear
nn.Conv2d(hidden_dim, oup, 1, 1, 0, bias=False),
nn.BatchNorm2d(oup),
)
def forward(self, x):
if self.use_res_connect:
return x + self.conv(x)
else:
return self.conv(x)
class MobileViTBlock(nn.Module):
def __init__(self, dim, depth, channel, kernel_size, patch_size, mlp_dim, dropout=0.):
super().__init__()
self.ph, self.pw = patch_size
self.conv1 = conv_nxn_bn(channel, channel, kernel_size)
self.conv2 = conv_1x1_bn(channel, dim)
self.transformer = Transformer(dim, depth, 4, 8, mlp_dim, dropout)
self.conv3 = conv_1x1_bn(dim, channel)
self.conv4 = conv_nxn_bn(2 * channel, channel, kernel_size)
def forward(self, x):
y = x.clone()
# Local representations
x = self.conv1(x)
x = self.conv2(x)
# Global representations
_, _, h, w = x.shape
x = rearrange(x, 'b d (h ph) (w pw) -> b (ph pw) (h w) d', ph=self.ph, pw=self.pw)
x = self.transformer(x)
x = rearrange(x, 'b (ph pw) (h w) d -> b d (h ph) (w pw)', h=h//self.ph, w=w//self.pw, ph=self.ph, pw=self.pw)
# Fusion
x = self.conv3(x)
x = torch.cat((x, y), 1)
x = self.conv4(x)
return x
class MobileViT(nn.Module):
def __init__(self, image_size, dims, channels, num_classes, expansion=4, kernel_size=3, patch_size=(2, 2)):
super().__init__()
ih, iw = image_size
ph, pw = patch_size
assert ih % ph == 0 and iw % pw == 0
L = [2, 4, 3]
self.conv1 = conv_nxn_bn(3, channels[0], stride=2)
self.mv2 = nn.ModuleList([])
self.mv2.append(MV2Block(channels[0], channels[1], 1, expansion))
self.mv2.append(MV2Block(channels[1], channels[2], 2, expansion))
self.mv2.append(MV2Block(channels[2], channels[3], 1, expansion))
self.mv2.append(MV2Block(channels[2], channels[3], 1, expansion)) # Repeat
self.mv2.append(MV2Block(channels[3], channels[4], 2, expansion))
self.mv2.append(MV2Block(channels[5], channels[6], 2, expansion))
self.mv2.append(MV2Block(channels[7], channels[8], 2, expansion))
self.mvit = nn.ModuleList([])
self.mvit.append(MobileViTBlock(dims[0], L[0], channels[5], kernel_size, patch_size, int(dims[0]*2)))
self.mvit.append(MobileViTBlock(dims[1], L[1], channels[7], kernel_size, patch_size, int(dims[1]*4)))
self.mvit.append(MobileViTBlock(dims[2], L[2], channels[9], kernel_size, patch_size, int(dims[2]*4)))
self.conv2 = conv_1x1_bn(channels[-2], channels[-1])
self.pool = nn.AvgPool2d(ih//32, 1)
self.fc = nn.Linear(channels[-1], num_classes, bias=False)
def forward(self, x):
x = self.conv1(x)
x = self.mv2[0](x)
x = self.mv2[1](x)
x = self.mv2[2](x)
x = self.mv2[3](x) # Repeat
x = self.mv2[4](x)
x = self.mvit[0](x)
x = self.mv2[5](x)
x = self.mvit[1](x)
x = self.mv2[6](x)
x = self.mvit[2](x)
x = self.conv2(x)
x = self.pool(x).view(-1, x.shape[1])
x = self.fc(x)
return x
def mobilevit_xxs(img_size=(256, 256), num_classes=1000):
dims = [64, 80, 96]
channels = [16, 16, 24, 24, 48, 48, 64, 64, 80, 80, 320]
return MobileViT((img_size[0], img_size[1]), dims, channels, num_classes=num_classes, expansion=2)
def mobilevit_xs(img_size=(256, 256), num_classes=1000):
dims = [96, 120, 144]
channels = [16, 32, 48, 48, 64, 64, 80, 80, 96, 96, 384]
return MobileViT((img_size[0], img_size[1]), dims, channels, num_classes=num_classes)
def mobilevit_s(img_size=(256, 256), num_classes=1000):
dims = [144, 192, 240]
channels = [16, 32, 64, 64, 96, 96, 128, 128, 160, 160, 640]
return MobileViT((img_size[0], img_size[1]), dims, channels, num_classes=num_classes)
def count_parameters(model):
return sum(p.numel() for p in model.parameters() if p.requires_grad)
if __name__ == '__main__':
img = torch.randn(5, 3, 256, 256)
vit = mobilevit_xxs(img_size=(256, 256))
out = vit(img)
print(out.shape)
print(count_parameters(vit))
vit = mobilevit_xs()
out = vit(img)
print(out.shape)
print(count_parameters(vit))
vit = mobilevit_s()
out = vit(img)
print(out.shape)
print(count_parameters(vit))