nsga2 matlab,NSGA2算法特征选择MATLAB实现(多目标)
利用nsga2进行进行特征选择其主要思想是:将子集的选择看作是一个搜索寻优问题(wrapper方法),生成不同的组合,对组合进行评价,再与其他的组合进行比较。这样就将子集的选择看作是一个是一个优化问题。
需要优化的两个目标为特征数和精度。
nsga2是一个多目标优化算法。
具体的特征选择代码在上述代码的基础上改了两个①主函数②评价函数,增加了一个数据分成训练集和测试集的函数:
MATLAB
function divide_datasets()
load Parkinson.mat;
dataMat=Parkinson_f;
len=size(dataMat,1);
%归一化
maxV = max(dataMat);
minV = min(dataMat);
range = maxV-minV;
newdataMat = (dataMat-repmat(minV,[len,1]))./(repmat(range,[len,1]));
Indices = crossvalind('Kfold', length(Parkinson_label), 10);
site = find(Indices==1|Indices==2|Indices==3);
train_F = newdataMat(site,:);
train_L = Parkinson_label(site);
site2 = find(Indices~=1&Indices~=2&Indices~=3);
test_F = newdataMat(site2,:);
test_L =Parkinson_label(site2);
save train_F train_F;
save train_L train_L;
save test_F test_F;
save test_L test_L;
end
%what doesn't kill you makes you stronger, stand a little taller,doesn't mean i'm over cause you're gonw.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
functiondivide_datasets()
loadParkinson.mat;
dataMat=Parkinson_f;
len=size(dataMat,1);
%归一化
maxV=max(dataMat);
minV=min(dataMat);
range=maxV-minV;
newdataMat=(dataMat-repmat(minV,[len,1]))./(repmat(range,[len,1]));
Indices=crossvalind('Kfold',length(Parkinson_label),10);
site=find(Indices==1|Indices==2|Indices==3);
train_F=newdataMat(site,:);
train_L=Parkinson_label(site);
site2=find(Indices~=1&Indices~=2&Indices~=3);
test_F=newdataMat(site2,:);
test_L=Parkinson_label(site2);
savetrain_Ftrain_F;
savetrain_Ltrain_L;
savetest_Ftest_F;
savetest_Ltest_L;
end
%what doesn't kill you makes you stronger, stand a little taller,doesn't mean i'm over cause you're gonw.
MATLAB代码主函数:
MATLAB
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%此处可以更改
%更多机器学习内容请访问omegaxyz.com
clc;
clear;
pop = 500; %种群数量
gen = 100; %迭代次数
M = 2; %目标数量
V = 22; %维度
min_range = zeros(1, V); %下界
max_range = ones(1,V); %上界
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%特征选择
divide_datasets();
global answer
answer=cell(M,3);
global choice %选出的特征个数
choice=0.8;
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
chromosome = initialize_variables(pop, M, V, min_range, max_range);
chromosome = non_domination_sort_mod(chromosome, M, V);
for i = 1 : gen
pool = round(pop/2);
tour = 2;
parent_chromosome = tournament_selection(chromosome, pool, tour);
mu = 20;
mum = 20;
offspring_chromosome = genetic_operator(parent_chromosome,M, V, mu, mum, min_range, max_range);
[main_pop,~] = size(chromosome);
[offspring_pop,~] = size(offspring_chromosome);
clear temp
intermediate_chromosome(1:main_pop,:) = chromosome;
intermediate_chromosome(main_pop + 1 : main_pop + offspring_pop,1 : M+V) = offspring_chromosome;
intermediate_chromosome = non_domination_sort_mod(intermediate_chromosome, M, V);
chromosome = replace_chromosome(intermediate_chromosome, M, V, pop);
if ~mod(i,100)
clc;
fprintf('%d generations completed\n',i);
end
end
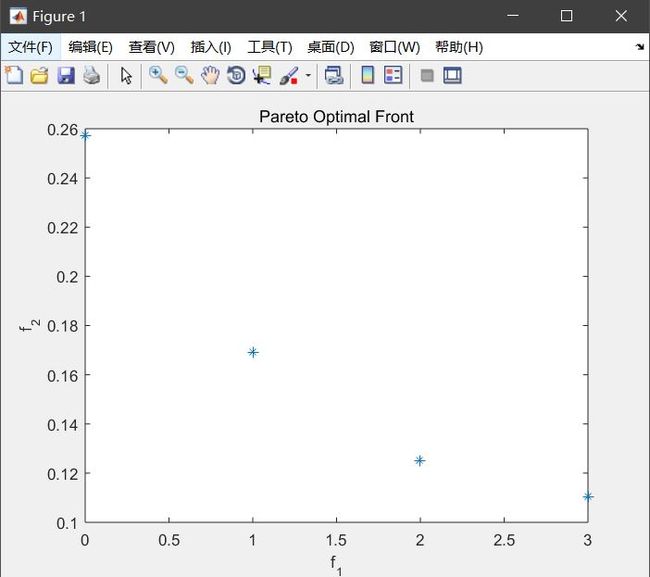
if M == 2
plot(chromosome(:,V + 1),chromosome(:,V + 2),'*');
xlabel('f_1'); ylabel('f_2');
title('Pareto Optimal Front');
elseif M == 3
plot3(chromosome(:,V + 1),chromosome(:,V + 2),chromosome(:,V + 3),'*');
xlabel('f_1'); ylabel('f_2'); zlabel('f_3');
title('Pareto Optimal Surface');
end
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%此处可以更改
%更多机器学习内容请访问omegaxyz.com
clc;
clear;
pop=500;%种群数量
gen=100;%迭代次数
M=2;%目标数量
V=22;%维度
min_range=zeros(1,V);%下界
max_range=ones(1,V);%上界
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%特征选择
divide_datasets();
globalanswer
answer=cell(M,3);
globalchoice%选出的特征个数
choice=0.8;
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
chromosome=initialize_variables(pop,M,V,min_range,max_range);
chromosome=non_domination_sort_mod(chromosome,M,V);
fori=1:gen
pool=round(pop/2);
tour=2;
parent_chromosome=tournament_selection(chromosome,pool,tour);
mu=20;
mum=20;
offspring_chromosome=genetic_operator(parent_chromosome,M,V,mu,mum,min_range,max_range);
[main_pop,~]=size(chromosome);
[offspring_pop,~]=size(offspring_chromosome);
cleartemp
intermediate_chromosome(1:main_pop,:)=chromosome;
intermediate_chromosome(main_pop+1:main_pop+offspring_pop,1:M+V)=offspring_chromosome;
intermediate_chromosome=non_domination_sort_mod(intermediate_chromosome,M,V);
chromosome=replace_chromosome(intermediate_chromosome,M,V,pop);
if~mod(i,100)
clc;
fprintf('%d generations completed\n',i);
end
end
ifM==2
plot(chromosome(:,V+1),chromosome(:,V+2),'*');
xlabel('f_1');ylabel('f_2');
title('Pareto Optimal Front');
elseifM==3
plot3(chromosome(:,V+1),chromosome(:,V+2),chromosome(:,V+3),'*');
xlabel('f_1');ylabel('f_2');zlabel('f_3');
title('Pareto Optimal Surface');
end
评价函数(利用林志仁SVM进行训练):
MATLAB
function f = evaluate_objective(x, M, V, i)
f = [];
global answer
global choice
load train_F.mat;
load train_L.mat;
load test_F.mat;
load test_L.mat;
temp_x = x(1:V);
inmodel = temp_x>choice;%%%%%设定恰当的阈值选择特征
f(1) = sum(inmodel(1,:));
answer(i,1)={f(1)};
model = libsvmtrain(train_L,train_F(:,inmodel), '-s 0 -t 2 -c 1.2 -g 2.8');
[predict_label, ~, ~] = libsvmpredict(test_L,test_F(:,inmodel),model,'-q');
error=0;
for j=1:length(test_L)
if(predict_label(j,1) ~= test_L(j,1))
error = error+1;
end
end
error = error/length(test_L);
f(2) = error;
answer(i,2)={error};
answer(i,3)={inmodel};
end
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
functionf=evaluate_objective(x,M,V,i)
f=[];
globalanswer
globalchoice
loadtrain_F.mat;
loadtrain_L.mat;
loadtest_F.mat;
loadtest_L.mat;
temp_x=x(1:V);
inmodel=temp_x>choice;%%%%%设定恰当的阈值选择特征
f(1)=sum(inmodel(1,:));
answer(i,1)={f(1)};
model=libsvmtrain(train_L,train_F(:,inmodel),'-s 0 -t 2 -c 1.2 -g 2.8');
[predict_label,~,~]=libsvmpredict(test_L,test_F(:,inmodel),model,'-q');
error=0;
forj=1:length(test_L)
if(predict_label(j,1)~=test_L(j,1))
error=error+1;
end
end
error=error/length(test_L);
f(2)=error;
answer(i,2)={error};
answer(i,3)={inmodel};
end
选的的数据集请从UCI上下载。
结果:
①pareto面
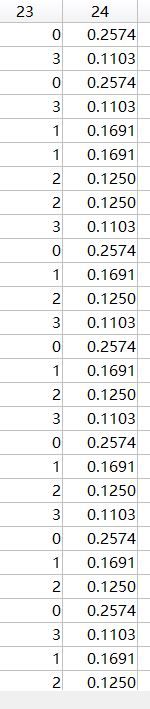
最后粒子的数据(选出的特征数和精确度)