nnUnet使用2d数据训练方法-DKFZ官方版
nnUnet使用2d数据训练方法-DKFZ官方版
上一篇文章介绍了《保姆级教程:nnUnet在2维图像的训练和测试》,采用的是自己的2d数据集进行2d到3d的数据转换,内容包括nnUnet介绍、环境配置、数据配置、预处理、训练过程、确定最佳的U-Net配置、运行推断,算是带着大家在2d数据情况下把nnUnet训练和测试了一遍。
最近官方也更新了nnUnet在2d数据情况下的训练方法,链接为:https://github.com/MIC-DKFZ/nnUNet/blob/master/documentation/dataset_conversion.md,将2d的马萨诸塞州道路数据road_segmentation_ideal转换成3d的Task120_MassRoadsSeg数据。
马萨诸塞州道路数据集是卫星标注图像,从航空图像中分割道路是一项具有挑战性的任务。来自附近树木的障碍物、相邻建筑物的阴影、道路纹理和颜色的变化、道路等级的不平衡(由于道路图像像素相对较少)是阻碍当前模型分割从图像一端延伸到另一端的尖锐道路边界的其他挑战。高质量的航空图像数据集有助于对现有方法进行比较,并在机器学习和计算机视觉领域引起人们对航空图像应用的兴趣。
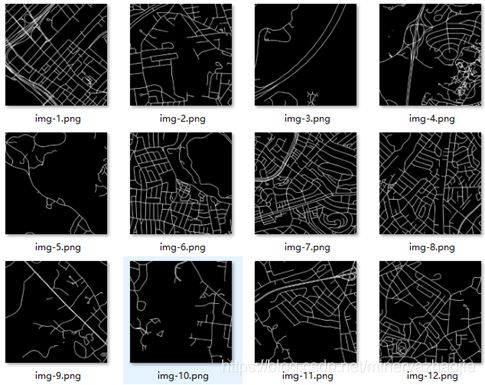
输入图像如下:

标注图像如下:
一、官方说明
How to use 2D data with nnU-Net
nnU-Net was originally built for 3D images. It is also strongestwhen applied to 3D segmentation problems because a large proportion of itsdesign choices were built with 3D in mind. Also note that many 2D segmentationproblems, especially in the non-biomedical domain, may benefit from pretrainednetwork architectures which nnU-Net does not support. Still, there is certainlya need for an out of the box segmentation solution for 2D segmentationproblems. And also on 2D segmentation tasks nnU-Net cam perform extremely well!We have, for example, won a 2D task in the cell tracking challenge with nnU-Net(see our Nature Methods paper) and we have also successfully applied nnU-Net tohistopathological segmentation problems. Working with 2D data in nnU-Netrequires a small workaround in the creation of the dataset. Essentially, allimages must be converted to pseudo 3D images (so an image with shape (X, Y)needs to be converted to an image with shape (1, X, Y). The resulting imagemust be saved in nifti format. Hereby it is important to set the spacing of thefirst axis (the one with shape 1) to a value larger than the others. If you areworking with niftis anyways, then doing this should be easy for you. Thisexample here is intended for demonstrating how nnU-Net can be used with’regular’ 2D images. We selected the massachusetts road segmentation datasetfor this because it can be obtained easily, it comes with a good amount oftraining cases but is still not too large to be difficult to handle.
See here for anexample. This script contains a lot of comments and useful information. Alsohave a look here.
How to convert other image formats tonifti
Please have a look at the following tasks:
Task120: 2D png images
Task075 and Task076: 3D tiff
Task089 2D tiff
二、Task120数据集
这里Task120的链接为:
https://github.com/MIC-DKFZ/nnUNet/blob/master/nnunet/dataset_conversion/Task120_Massachusetts_RoadSegm.py
首先简单介绍一下代码的功能:
-
创建原始数据的文件夹Task120_MassRoadsSeg以及子文件夹imagesTr、imagesTs、labelsTs、labelsTr。
-
/road_segmentation_ideal路径内的有训练集training和测试集testing两个文件夹,数据集文件夹内有输入图像input和标签output两个文件夹,分别提取到每个数据的名称。
-
调用convert_2d_image_to_nifti函数,读取每个数据,将3通道的图像拆成3个模态的3个数据,设置每个像素点实际的长度与宽度Spacing,并保存成nii.gz的3维数据,这里的3维数据其实只有1层。
原始图像img-1.png如下:
生成的3个通道的3个模态数据img-1_0000.nii.gz、img-1_0001.nii.gz和img-1_0002.nii.gz如下,是用ITK-SNAP打开的,只展示沿z轴的一层图像。

三、代码详解
此部分代码文件为:
nnUNet/nnunet/dataset_conversion/Task120_Massachusetts_RoadSegm.py
1. 首先导入需要的包
import numpy as np
from batchgenerators.utilities.file_and_folder_operations import *
from nnunet.dataset_conversion.utils import generate_dataset_json
from nnunet.paths import nnUNet_raw_data, preprocessing_output_dir
from nnunet.utilities.file_conversions import convert_2d_image_to_nifti
2. 定义main函数并做介绍
if __name__ == '__main__':
"""
nnU-Net was originally built for 3D images. It is also strongest when applied to 3D segmentation problems because a
large proportion of its design choices were built with 3D in mind. Also note that many 2D segmentation problems,
especially in the non-biomedical domain, may benefit from pretrained network architectures which nnU-Net does not
support.
Still, there is certainly a need for an out of the box segmentation solution for 2D segmentation problems. And
also on 2D segmentation tasks nnU-Net cam perform extremely well! We have, for example, won a 2D task in the cell
tracking challenge with nnU-Net (see our Nature Methods paper) and we have also successfully applied nnU-Net to
histopathological segmentation problems.
Working with 2D data in nnU-Net requires a small workaround in the creation of the dataset. Essentially, all images
must be converted to pseudo 3D images (so an image with shape (X, Y) needs to be converted to an image with shape
(1, X, Y). The resulting image must be saved in nifti format. Hereby it is important to set the spacing of the
first axis (the one with shape 1) to a value larger than the others. If you are working with niftis anyways, then
doing this should be easy for you. This example here is intended for demonstrating how nnU-Net can be used with
'regular' 2D images. We selected the massachusetts road segmentation dataset for this because it can be obtained
easily, it comes with a good amount of training cases but is still not too large to be difficult to handle.
"""
大致意思是nnUnet最初适用于3D数据,当处理2D数据时需要将数据做成一个伪3D的图像,形状为(X,Y)的图像需要转换为形状为(1,X,Y)的图像,结果图像必须以nifti格式保存,将第一轴(具有形状1的轴)的间距设置为大于其他轴的值。该数据集为2d数据马萨诸塞州道路分割数据集。
3. 下载数据集、修改文件路径并创建nnUnet能识别的文件夹
# download dataset from https://www.kaggle.com/insaff/massachusetts-roads-dataset
# extract the zip file, then set the following path according to your system:
base = '/media/fabian/data/road_segmentation_ideal'
# this folder should have the training and testing subfolders
# now start the conversion to nnU-Net:
task_name = 'Task120_MassRoadsSeg'
target_base = join(nnUNet_raw_data, task_name)
target_imagesTr = join(target_base, "imagesTr")
target_imagesTs = join(target_base, "imagesTs")
target_labelsTs = join(target_base, "labelsTs")
target_labelsTr = join(target_base, "labelsTr")
maybe_mkdir_p(target_imagesTr)
maybe_mkdir_p(target_labelsTs)
maybe_mkdir_p(target_imagesTs)
maybe_mkdir_p(target_labelsTr)
这里给出了数据集的下载链接:
https://www.kaggle.com/insaff/massachusetts-roads-dataset,解压并修改文件路径base。
创建原始数据的文件夹Task120_MassRoadsSeg以及子文件夹imagesTr、imagesTs、labelsTs、labelsTr。
4. 对训练数据文件进行遍历
# convert the training examples. Not all training images have labels, so we just take the cases for which there are
# labels
labels_dir_tr = join(base, 'training', 'output')
images_dir_tr = join(base, 'training', 'input')
training_cases = subfiles(labels_dir_tr, suffix='.png', join=False)
for t in training_cases:
unique_name = t[:-4] # just the filename with the extension cropped away, so img-2.png becomes img-2 as unique_name
input_segmentation_file = join(labels_dir_tr, t)
input_image_file = join(images_dir_tr, t)
output_image_file = join(target_imagesTr, unique_name) # do not specify a file ending! This will be done for you
output_seg_file = join(target_labelsTr, unique_name) # do not specify a file ending! This will be done for you
这里提示一下,该数据集部分数据无标签,所以代码查找的数据以有label的数据为准,无label的数据直接忽略。
针对/road_segmentation_ideal路径内的训练集training文件夹,数据集文件夹内有输入图像input和标签output两个文件夹,输入图像文件夹为images_dir_tr,标签图像文件夹为labels_dir_tr,提取到output文件夹里的所有数据名称training_cases。
分别依次读取每个数据(这里是3通道的彩色2d图像),获取数据名称unique_name(去掉后缀),输入图像文件为input_image_file,标注图像文件为input_segmentation_file,输出图像文件为output_image_file(不带后缀,后面会分成3个模态3个输出图像文件),输出标注文件output_seg_file。
5. 调用convert_2d_image_to_nifti函数进行数据转换
# this utility will convert 2d images that can be read by skimage.io.imread to nifti. You don't need to do anything.
# if this throws an error for your images, please just look at the code for this function and adapt it to your needs
convert_2d_image_to_nifti(input_image_file, output_image_file, is_seg=False)
# the labels are stored as 0: background, 255: road. We need to convert the 255 to 1 because nnU-Net expects
# the labels to be consecutive integers. This can be achieved with setting a transform
convert_2d_image_to_nifti(input_segmentation_file, output_seg_file, is_seg=True,
transform=lambda x: (x == 255).astype(int))
上面的代码是对训练数据的输入图像和标签图像做转换,调用了convert_2d_image_to_nifti函数,该函数首先做了定义和一些介绍:
from typing import Tuple, List
from skimage import io
import SimpleITK as sitk
import numpy as np
import tifffile
def convert_2d_image_to_nifti(input_filename: str, output_filename_truncated: str, spacing=(999, 1, 1),
transform=None, is_seg: bool = False) -> None:
"""
Reads an image (must be a format that it recognized by skimage.io.imread) and converts it into a series of niftis.
The image can have an arbitrary number of input channels which will be exported separately (_0000.nii.gz,
_0001.nii.gz, etc for images and only .nii.gz for seg).
Spacing can be ignored most of the time.
!!!2D images are often natural images which do not have a voxel spacing that could be used for resampling. These images
must be resampled by you prior to converting them to nifti!!!
Datasets converted with this utility can only be used with the 2d U-Net configuration of nnU-Net
If Transform is not None it will be applied to the image after loading.
Segmentations will be converted to np.uint32!
:param is_seg:
:param transform:
:param input_filename:
:param output_filename_truncated: do not use a file ending for this one! Example: output_name='./converted/image1'. This
function will add the suffix (_0000) and file ending (.nii.gz) for you.
:param spacing:
:return:
"""
大致意思是要做数据转换了,输入图像可以是任意通道的(通常是3通道),后面会单独输出_0000.nii.gz、_0001.nii.gz等的结果,但标签数据还是输出不带模态标志的.nii.gz,间距spacing可以忽略,转换之后的数据集只能用2d unet配置使用。
首先用skimage读图
img = io.imread(input_filename)
如果是标签图像,则应用transform,将[0,255]的标签图像转换成[0,1]的标签图像
if transform is not None:
img = transform(img)
这里的
transform=lambda x: (x == 255).astype(int))
如果是1通道的2d图像,则图像形状可能是(256,256),需要增加2个维度,变成(1,1,256,256)
if len(img.shape) == 2: # 2d image with no color channels
img = img[None, None] # add dimensions
如果是3通道的2d图像,则图像形状可能是(256,256,3),先转置成(3,256,256),然后增加1个维度,变成(3, 1, 256, 256)。
else:
assert len(img.shape) == 3, "image should be 3d with color channel last but has shape %s" % str(img.shape)
# we assume that the color channel is the last dimension. Transpose it to be in first
img = img.transpose((2, 0, 1))
# add third dimension
img = img[:, None]
再次确认标签图像的通道数为1
# image is now (c, x, x, z) where x=1 since it's 2d
if is_seg:
assert img.shape[0] == 1, 'segmentations can only have one color channel, not sure what happened here'
遍历图像的通道数(通常为3个),分别提取不同通道的图像,转换成itk文件,设置spacing层厚,这里预定义的spacing=(999, 1, 1),经过list(spacing)[::-1]后变成[1, 1, 999],sitk图像顺序是x,y,z三个方向的大小,numpy矩阵的顺序是z,y,x三个方向的大小,所以spacing需要转换成[1, 1, 999]。之后针对输入图像和标签图像分别存储为带模态标志0000/0001/0002的nifti数据和不带模态标志的nifti数据。
for j, i in enumerate(img):
if is_seg:
i = i.astype(np.uint32)
itk_img = sitk.GetImageFromArray(i)
itk_img.SetSpacing(list(spacing)[::-1])
if not is_seg:
sitk.WriteImage(itk_img, output_filename_truncated + "_%04.0d.nii.gz" % j)
else:
sitk.WriteImage(itk_img, output_filename_truncated + ".nii.gz")
测试数据方法一样,
# now do the same for the test set
labels_dir_ts = join(base, 'testing', 'output')
images_dir_ts = join(base, 'testing', 'input')
testing_cases = subfiles(labels_dir_ts, suffix='.png', join=False)
for ts in testing_cases:
unique_name = ts[:-4]
input_segmentation_file = join(labels_dir_ts, ts)
input_image_file = join(images_dir_ts, ts)
output_image_file = join(target_imagesTs, unique_name)
output_seg_file = join(target_labelsTs, unique_name)
convert_2d_image_to_nifti(input_image_file, output_image_file, is_seg=False)
convert_2d_image_to_nifti(input_segmentation_file, output_seg_file, is_seg=True,
transform=lambda x: (x == 255).astype(int))
最后生成json文件
# finally we can call the utility for generating a dataset.json
generate_dataset_json(join(target_base, 'dataset.json'), target_imagesTr, target_imagesTs, ('Red', 'Green', 'Blue'),
labels={1: 'street'}, dataset_name=task_name, license='hands off!')
最终转换之后的数据格式为:
nnUNet_raw_data_base/nnUNet_raw_data/Task120_MassRoadsSeg
├── dataset.json
├── imagesTr
│ ├── img-2_0000.nii.gz
│ ├── img-2_0001.nii.gz
│ ├── img-2_0002.nii.gz
│ ├── ...
├── imagesTs
│ ├── img-1_0000.nii.gz
│ ├── img-1_0001.nii.gz
│ ├── img-1_0002.nii.gz
│ ├── ...
├── labelsTr
│ ├── img-2.nii.gz
│ ├── img-7.nii.gz
│ ├── ...
├── labelsTs
│ ├── img-1.nii.gz
│ ├── img-2.nii.gz
│ ├── ...
这样数据集就处理好了,之后再按照步骤进行预处理、训练过程、确定最佳的U-Net配置、运行推断就好了,可以参照官方https://github.com/MIC-DKFZ/nnUNet,也可以参考我们的前一篇文章《保姆级教程:nnUnet在2维图像的训练和测试》。
最后2021年冲鸭!!!
原文链接:https://mp.weixin.qq.com/s?__biz=MzUxNTY1MjMxNQ==&mid=2247485775&idx=1&sn=cbf734d32b33bc8d80f24fbabebedef8&chksm=f9b226fbcec5afed3efec9404ae4c7691b9ea445a9a360777c3d424529ceba6cbbf24a5dbf92&token=409175063&lang=zh_CN#rd