tidymodels搞定二分类资料多个模型评价和比较
本文首发于公众号:医学和生信笔记
“医学和生信笔记,专注R语言在临床医学中的使用,R语言数据分析和可视化。主要分享R语言做医学统计学、meta分析、网络药理学、临床预测模型、机器学习、生物信息学等。
前面介绍了很多二分类资料的模型评价内容,用到了很多R包,虽然达到了目的,但是内容太多了,不太容易记住。
今天给大家介绍一个很厉害的R包:tidymodels,一个R包搞定二分类资料的模型评价和比较。
一看这个名字就知道,和tidyverse系列师出同门,包的作者是大佬Max Kuhn,大佬的上一个作品是caret,现在加盟rstudio了,开发了新的机器学习R包,也就是今天要介绍的tidymodels。
给大家看看如何用优雅的方式建立、评价、比较多个模型!
本期目录:
- 加载数据和R包
- 数据划分
- 数据预处理
- 建立多个模型
- logistic
- knn
- 随机森林
- 决策树
- 交叉验证
- ROC曲线画一起
加载数据和R包
没有安装的R包的自己安装下~
suppressPackageStartupMessages(library(tidyverse))
suppressPackageStartupMessages(library(tidymodels))
tidymodels_prefer()
由于要做演示用,肯定要一份比较好的数据才能说明问题,今天用的这份数据,结果变量是一个二分类的。
一共有91976行,26列,其中play_type是结果变量,因子型,其余列都是预测变量。
all_plays <- read_rds("../000files/all_plays.rds")
glimpse(all_plays)
## Rows: 91,976
## Columns: 26
## $ game_id 2017090700, 2017090700, 2017090700, 2017090…
## $ posteam "NE", "NE", "NE", "NE", "NE", "NE", "NE", "…
## $ play_type pass, pass, run, run, pass, run, pass, pass…
## $ yards_gained 0, 8, 8, 3, 19, 5, 16, 0, 2, 7, 0, 3, 10, 0…
## $ ydstogo 10, 10, 2, 10, 7, 10, 5, 2, 2, 10, 10, 10, …
## $ down 1, 2, 3, 1, 2, 1, 2, 1, 2, 1, 1, 2, 3, 1, 2…
## $ game_seconds_remaining 3595, 3589, 3554, 3532, 3506, 3482, 3455, 3…
## $ yardline_100 73, 73, 65, 57, 54, 35, 30, 2, 2, 75, 32, 3…
## $ qtr 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1…
## $ posteam_score 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 7, 7, 7, 7, 7…
## $ defteam "KC", "KC", "KC", "KC", "KC", "KC", "KC", "…
## $ defteam_score 0, 0, 0, 0, 0, 0, 0, 0, 0, 7, 0, 0, 0, 0, 0…
## $ score_differential 0, 0, 0, 0, 0, 0, 0, 0, 0, -7, 7, 7, 7, 7, …
## $ shotgun 0, 0, 1, 1, 1, 0, 1, 0, 0, 1, 1, 1, 1, 1, 0…
## $ no_huddle 0, 0, 0, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0…
## $ posteam_timeouts_remaining 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3…
## $ defteam_timeouts_remaining 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3…
## $ wp 0.5060180, 0.4840546, 0.5100098, 0.5529816,…
## $ goal_to_go 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0…
## $ half_seconds_remaining 1795, 1789, 1754, 1732, 1706, 1682, 1655, 1…
## $ total_runs 0, 0, 0, 1, 2, 2, 3, 3, 3, 0, 4, 4, 4, 5, 5…
## $ total_pass 0, 1, 2, 2, 2, 3, 3, 4, 5, 0, 5, 6, 7, 7, 8…
## $ previous_play First play of Drive, pass, pass, run, run, …
## $ in_red_zone 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 1, 1…
## $ in_fg_range 0, 0, 0, 0, 0, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1…
## $ two_min_drill 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
数据划分
把75%的数据用于训练集,剩下的做测试集。
set.seed(20220520)
# 数据划分,根据play_type分层
split_pbp <- initial_split(all_plays, 0.75, strata = play_type)
train_data <- training(split_pbp) # 训练集
test_data <- testing(split_pbp) # 测试集
数据预处理
pbp_rec <- recipe(play_type ~ ., data = train_data) %>%
step_rm(half_seconds_remaining,yards_gained, game_id) %>% # 移除这3列
step_string2factor(posteam, defteam) %>% # 变为因子类型
#update_role(yards_gained, game_id, new_role = "ID") %>%
# 去掉高度相关的变量
step_corr(all_numeric(), threshold = 0.7) %>%
step_center(all_numeric()) %>% # 中心化
step_zv(all_predictors()) # 去掉零方差变量
建立多个模型
logistic
选择模型,连接数据预处理步骤。
lm_spec <- logistic_reg(mode = "classification",engine = "glm")
lm_wflow <- workflow() %>%
add_recipe(pbp_rec) %>%
add_model(lm_spec)
建立模型:
fit_lm <- lm_wflow %>% fit(data = train_data)
应用于测试集:
pred_lm <- select(test_data, play_type) %>%
bind_cols(predict(fit_lm, test_data, type = "prob")) %>%
bind_cols(predict(fit_lm, test_data))
查看模型表现:
# 选择多种评价指标
metricsets <- metric_set(accuracy, mcc, f_meas, j_index)
pred_lm %>% metricsets(truth = play_type, estimate = .pred_class)
## # A tibble: 4 × 3
## .metric .estimator .estimate
##
## 1 accuracy binary 0.724
## 2 mcc binary 0.423
## 3 f_meas binary 0.774
## 4 j_index binary 0.416
大家最喜欢的AUC:
pred_lm %>% roc_auc(truth = play_type, .pred_pass)
## # A tibble: 1 × 3
## .metric .estimator .estimate
##
## 1 roc_auc binary 0.781
可视化结果,首先是大家喜闻乐见的ROC曲线:
pred_lm %>% roc_curve(truth = play_type, .pred_pass) %>%
autoplot()
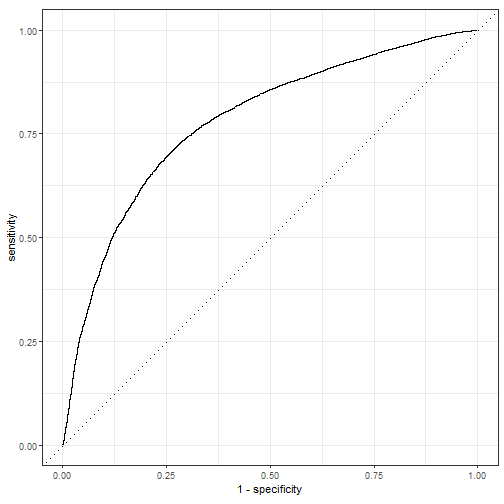 plot of chunk unnamed-chunk-10
plot of chunk unnamed-chunk-10
pr曲线:
pred_lm %>% pr_curve(truth = play_type, .pred_pass) %>%
autoplot()
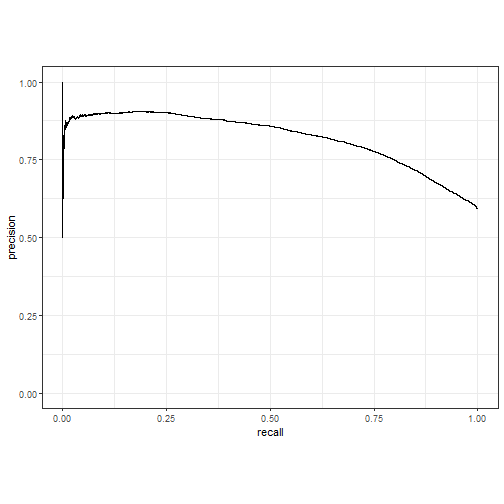 plot of chunk unnamed-chunk-11
plot of chunk unnamed-chunk-11
gain_curve:
pred_lm %>% gain_curve(truth = play_type, .pred_pass) %>%
autoplot()
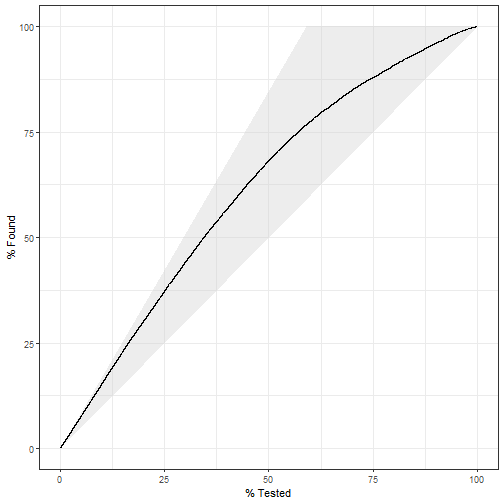 plot of chunk unnamed-chunk-12
plot of chunk unnamed-chunk-12
lift_curve:
pred_lm %>% lift_curve(truth = play_type, .pred_pass) %>%
autoplot()
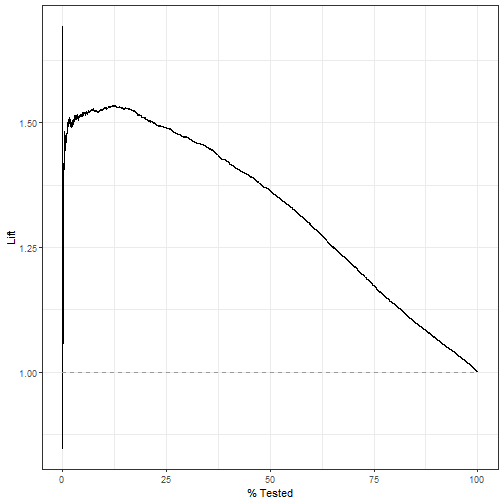 plot of chunk unnamed-chunk-13
plot of chunk unnamed-chunk-13
混淆矩阵:
pred_lm %>%
conf_mat(play_type,.pred_class) %>%
autoplot()
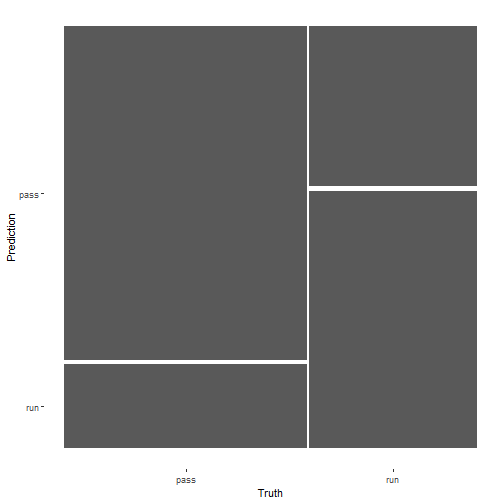 plot of chunk unnamed-chunk-14
plot of chunk unnamed-chunk-14
knn
k最近邻法,和上面的逻辑回归一模一样的流程。
首先也是选择模型,连接数据预处理步骤:
knn_spec <- nearest_neighbor(mode = "classification", engine = "kknn")
knn_wflow <- workflow() %>%
add_recipe(pbp_rec) %>%
add_model(knn_spec)
建立模型:
library(kknn)
fit_knn <- knn_wflow %>%
fit(train_data)
应用于测试集:
pred_knn <- test_data %>% select(play_type) %>%
bind_cols(predict(fit_knn, test_data, type = "prob")) %>%
bind_cols(predict(fit_knn, test_data, type = "class"))
查看模型表现:
metricsets <- metric_set(accuracy, mcc, f_meas, j_index)
pred_knn %>% metricsets(truth = play_type, estimate = .pred_class)
## # A tibble: 4 × 3
## .metric .estimator .estimate
##
## 1 accuracy binary 0.672
## 2 mcc binary 0.317
## 3 f_meas binary 0.727
## 4 j_index binary 0.315
pred_knn %>% roc_auc(play_type, .pred_pass)
## # A tibble: 1 × 3
## .metric .estimator .estimate
##
## 1 roc_auc binary 0.718
可视化模型的部分就不说了,和上面的一模一样!
随机森林
同样的流程来第3遍!
rf_spec <- rand_forest(mode = "classification") %>%
set_engine("ranger",importance = "permutation")
rf_wflow <- workflow() %>%
add_recipe(pbp_rec) %>%
add_model(rf_spec)
建立模型:
fit_rf <- rf_wflow %>%
fit(train_data)
应用于测试集:
pred_rf <- test_data %>% select(play_type) %>%
bind_cols(predict(fit_rf, test_data, type = "prob")) %>%
bind_cols(predict(fit_rf, test_data, type = "class"))
查看模型表现:
pred_rf %>% metricsets(truth = play_type, estimate = .pred_class)
## # A tibble: 4 × 3
## .metric .estimator .estimate
##
## 1 accuracy binary 0.731
## 2 mcc binary 0.441
## 3 f_meas binary 0.774
## 4 j_index binary 0.439
pred_rf %>% conf_mat(truth = play_type, estimate = .pred_class)
## Truth
## Prediction pass run
## pass 10622 3225
## run 2962 6186
pred_rf %>% roc_auc(play_type, .pred_pass)
## # A tibble: 1 × 3
## .metric .estimator .estimate
##
## 1 roc_auc binary 0.799
下面给大家手动画一个校准曲线。
两种画法,差别不大,主要是分组方法不一样,第2种分组方法是大家常见的哦~
calibration_df <- pred_rf %>%
mutate(pass = if_else(play_type == "pass", 1, 0),
pred_rnd = round(.pred_pass, 2)
) %>%
group_by(pred_rnd) %>%
summarize(mean_pred = mean(.pred_pass),
mean_obs = mean(pass),
n = n()
)
ggplot(calibration_df, aes(mean_pred, mean_obs))+
geom_point(aes(size = n), alpha = 0.5)+
geom_abline(linetype = "dashed")+
theme_minimal()
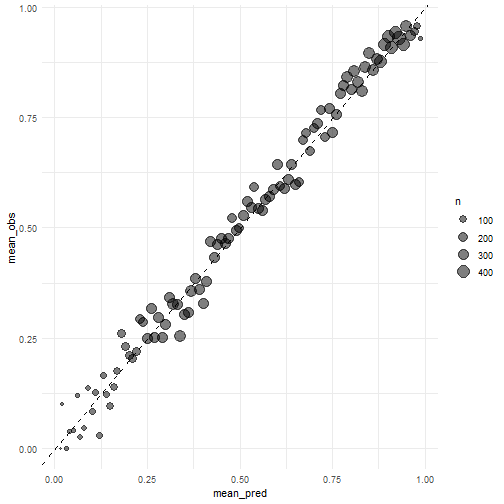 plot of chunk unnamed-chunk-26
plot of chunk unnamed-chunk-26
第2种方法:
cali_df <- pred_rf %>%
arrange(.pred_pass) %>%
mutate(pass = if_else(play_type == "pass", 1, 0),
group = c(rep(1:249,each=92), rep(250,87))
) %>%
group_by(group) %>%
summarise(mean_pred = mean(.pred_pass),
mean_obs = mean(pass)
)
cali_plot <- ggplot(cali_df, aes(mean_pred, mean_obs))+
geom_point(alpha = 0.5)+
geom_abline(linetype = "dashed")+
theme_minimal()
cali_plot
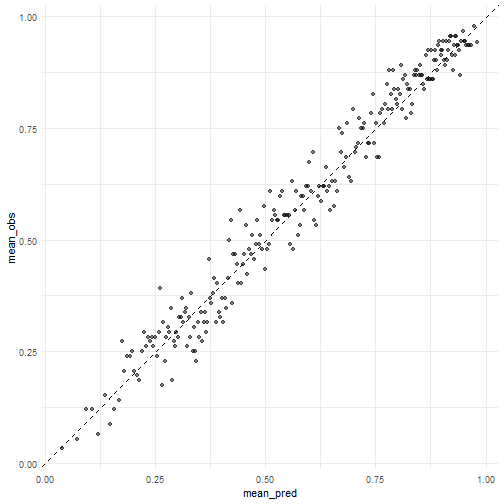 plot of chunk unnamed-chunk-27
plot of chunk unnamed-chunk-27
随机森林这种方法是可以计算变量重要性的,当然也是能把结果可视化的。
给大家演示下如何可视化随机森林结果的变量重要性:
library(vip)
fit_rf %>%
extract_fit_parsnip() %>%
vip(num_features = 10)
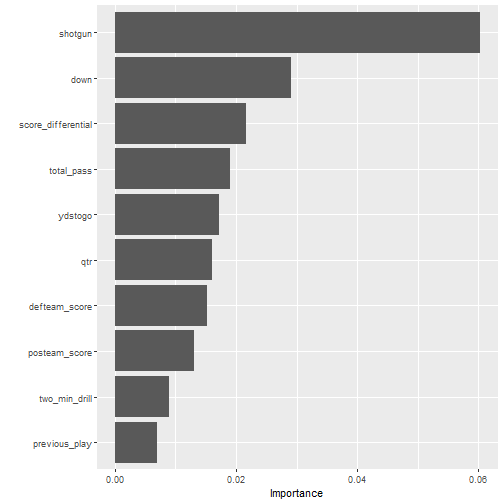 plot of chunk unnamed-chunk-28
plot of chunk unnamed-chunk-28
决策树
同样的流程来第4遍!不知道你看懂了没有。。。
tree_spec <- decision_tree(mode = "classification",engine = "rpart")
tree_wflow <- workflow() %>%
add_recipe(pbp_rec) %>%
add_model(tree_spec)
建立模型:
fit_tree <- tree_wflow %>%
fit(train_data)
应用于测试集:
pred_tree <- test_data %>% select(play_type) %>%
bind_cols(predict(fit_tree, test_data, type = "prob")) %>%
bind_cols(predict(fit_tree, test_data, type = "class"))
查看结果:
pred_tree %>% roc_auc(play_type, .pred_pass)
## # A tibble: 1 × 3
## .metric .estimator .estimate
##
## 1 roc_auc binary 0.706
pred_tree %>% metricsets(truth = play_type, estimate = .pred_class)
## # A tibble: 4 × 3
## .metric .estimator .estimate
##
## 1 accuracy binary 0.721
## 2 mcc binary 0.417
## 3 f_meas binary 0.770
## 4 j_index binary 0.411
交叉验证
交叉验证也是大家喜闻乐见的,就用随机森林给大家顺便演示下交叉验证。
首先要选择重抽样方法,这里我们选择10折交叉验证:
set.seed(20220520)
folds <- vfold_cv(train_data, v = 10)
folds
## # 10-fold cross-validation
## # A tibble: 10 × 2
## splits id
##
## 1 Fold01
## 2 Fold02
## 3 Fold03
## 4 Fold04
## 5 Fold05
## 6 Fold06
## 7 Fold07
## 8 Fold08
## 9 Fold09
## 10 Fold10
然后就是让模型在训练集上跑起来:
keep_pred <- control_resamples(save_pred = T, verbose = T)
set.seed(20220520)
library(doParallel)
## Loading required package: foreach
##
## Attaching package: 'foreach'
## The following objects are masked from 'package:purrr':
##
## accumulate, when
## Loading required package: iterators
## Loading required package: parallel
cl <- makePSOCKcluster(12) # 加速,用12个线程
registerDoParallel(cl)
rf_res <- fit_resamples(rf_wflow, resamples = folds, control = keep_pred)
i Fold01: preprocessor 1/1
✓ Fold01: preprocessor 1/1
i Fold01: preprocessor 1/1, model 1/1
✓ Fold01: preprocessor 1/1, model 1/1
i Fold01: preprocessor 1/1, model 1/1 (predictions)
i Fold02: preprocessor 1/1
✓ Fold02: preprocessor 1/1
i Fold02: preprocessor 1/1, model 1/1
✓ Fold02: preprocessor 1/1, model 1/1
i Fold02: preprocessor 1/1, model 1/1 (predictions)
i Fold03: preprocessor 1/1
✓ Fold03: preprocessor 1/1
i Fold03: preprocessor 1/1, model 1/1
✓ Fold03: preprocessor 1/1, model 1/1
i Fold03: preprocessor 1/1, model 1/1 (predictions)
i Fold04: preprocessor 1/1
✓ Fold04: preprocessor 1/1
i Fold04: preprocessor 1/1, model 1/1
✓ Fold04: preprocessor 1/1, model 1/1
i Fold04: preprocessor 1/1, model 1/1 (predictions)
i Fold05: preprocessor 1/1
✓ Fold05: preprocessor 1/1
i Fold05: preprocessor 1/1, model 1/1
✓ Fold05: preprocessor 1/1, model 1/1
i Fold05: preprocessor 1/1, model 1/1 (predictions)
i Fold06: preprocessor 1/1
✓ Fold06: preprocessor 1/1
i Fold06: preprocessor 1/1, model 1/1
✓ Fold06: preprocessor 1/1, model 1/1
i Fold06: preprocessor 1/1, model 1/1 (predictions)
i Fold07: preprocessor 1/1
✓ Fold07: preprocessor 1/1
i Fold07: preprocessor 1/1, model 1/1
✓ Fold07: preprocessor 1/1, model 1/1
i Fold07: preprocessor 1/1, model 1/1 (predictions)
i Fold08: preprocessor 1/1
✓ Fold08: preprocessor 1/1
i Fold08: preprocessor 1/1, model 1/1
✓ Fold08: preprocessor 1/1, model 1/1
i Fold08: preprocessor 1/1, model 1/1 (predictions)
i Fold09: preprocessor 1/1
✓ Fold09: preprocessor 1/1
i Fold09: preprocessor 1/1, model 1/1
✓ Fold09: preprocessor 1/1, model 1/1
i Fold09: preprocessor 1/1, model 1/1 (predictions)
i Fold10: preprocessor 1/1
✓ Fold10: preprocessor 1/1
i Fold10: preprocessor 1/1, model 1/1
✓ Fold10: preprocessor 1/1, model 1/1
i Fold10: preprocessor 1/1, model 1/1 (predictions)
stopCluster(cl)
查看模型表现:
rf_res %>%
collect_metrics(summarize = T)
## # A tibble: 2 × 6
## .metric .estimator mean n std_err .config
##
## 1 accuracy binary 0.732 10 0.00157 Preprocessor1_Model1
## 2 roc_auc binary 0.799 10 0.00193 Preprocessor1_Model1
查看具体的结果:
rf_res %>% collect_predictions()
## # A tibble: 68,981 × 7
## id .pred_pass .pred_run .row .pred_class play_type .config
##
## 1 Fold01 0.572 0.428 6 pass pass Preprocessor1_Model1
## 2 Fold01 0.470 0.530 8 run pass Preprocessor1_Model1
## 3 Fold01 0.898 0.102 22 pass pass Preprocessor1_Model1
## 4 Fold01 0.915 0.0847 69 pass pass Preprocessor1_Model1
## 5 Fold01 0.841 0.159 97 pass pass Preprocessor1_Model1
## 6 Fold01 0.931 0.0688 112 pass pass Preprocessor1_Model1
## 7 Fold01 0.729 0.271 123 pass pass Preprocessor1_Model1
## 8 Fold01 0.640 0.360 129 pass pass Preprocessor1_Model1
## 9 Fold01 0.740 0.260 136 pass pass Preprocessor1_Model1
## 10 Fold01 0.902 0.0979 143 pass pass Preprocessor1_Model1
## # … with 68,971 more rows
可视化结果也是和上面的一模一样,就不一一介绍了,简单说下训练集的校准曲线画法,其实也是和上面一样的~
res_calib_plot <- collect_predictions(rf_res) %>%
mutate(
pass = if_else(play_type == "pass", 1, 0),
pred_rnd = round(.pred_pass, 2)
) %>%
group_by(pred_rnd) %>%
summarize(
mean_pred = mean(.pred_pass),
mean_obs = mean(pass),
n = n()
) %>%
ggplot(aes(x = mean_pred, y = mean_obs)) +
geom_abline(linetype = "dashed") +
geom_point(aes(size = n), alpha = 0.5) +
theme_minimal() +
labs(
x = "Predicted Pass",
y = "Observed Pass"
) +
coord_cartesian(
xlim = c(0,1), ylim = c(0, 1)
)
res_calib_plot
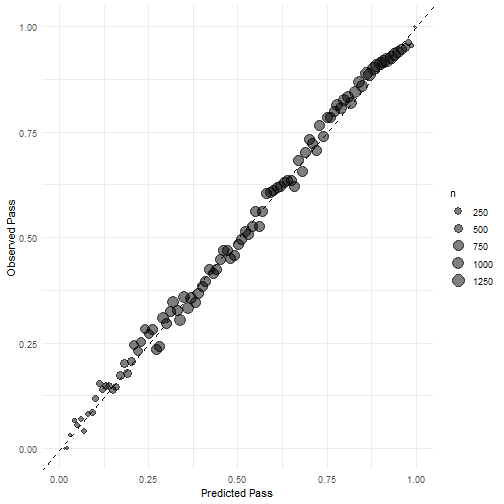 plot of chunk unnamed-chunk-38
plot of chunk unnamed-chunk-38
然后就是应用于测试集,并查看测试集上的表现:
rf_test_res <- last_fit(rf_wflow, split_pbp) %>%
collect_metrics()
## Error in summary.connection(connection): invalid connection
rf_test_res
# A tibble: 2 × 4
.metric .estimator .estimate .config
1 accuracy binary 0.730 Preprocessor1_Model1
2 roc_auc binary 0.798 Preprocessor1_Model1
ROC曲线画一起
其实非常简单,就是把结果拼在一起画个图就行了~
roc_lm <- pred_lm %>% roc_curve(play_type, .pred_pass) %>%
mutate(model = "logistic")
roc_knn <- pred_knn %>% roc_curve(play_type, .pred_pass) %>%
mutate(model = "kknn")
roc_rf <- pred_rf %>% roc_curve(play_type, .pred_pass) %>%
mutate(model = "randomforest")
roc_tree <- pred_tree %>% roc_curve(play_type, .pred_pass) %>%
mutate(model = "decision tree")
rocs <- bind_rows(roc_lm,roc_knn,roc_rf,roc_tree) %>%
ggplot(aes(x = 1 - specificity, y = sensitivity, color = model))+
geom_path(lwd = 1.2, alpha = 0.6)+
geom_abline(lty = 3)+
scale_color_brewer(palette = "Set1")+
theme_minimal()
rocs
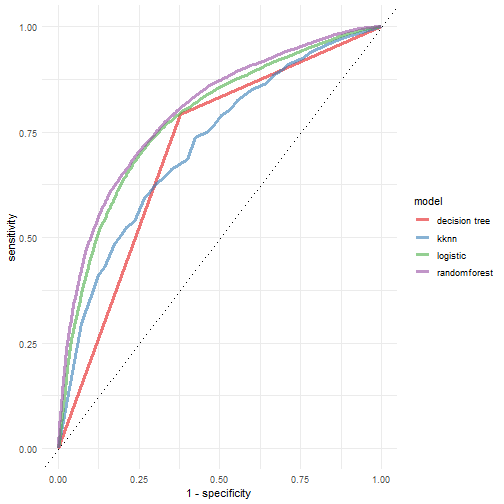 plot of chunk unnamed-chunk-41
plot of chunk unnamed-chunk-41
是不是很简单呢? 二分类资料常见的各种评价指标都有了,图也有了,还比较了多个模型,一举多得,tidymodels,你值得拥有!
本文首发于公众号:医学和生信笔记
“医学和生信笔记,专注R语言在临床医学中的使用,R语言数据分析和可视化。主要分享R语言做医学统计学、meta分析、网络药理学、临床预测模型、机器学习、生物信息学等。
本文由 mdnice 多平台发布