easylabel | 完美拯救手残党不会标注突出重点!(Label!~ Label!~)
1写在前面
我们在画图的时候经常需要标记某个值, 如散点图中的某个具体的点, 火山图中的某个基因, 但对于代码不太熟悉的小白来说, 还是有一定难度的.
本期和大家介绍一个基于shiny轻松进行label的包, 即easylabel包, 轻松实现交互式label, 麻麻再也不用担心你的画图标记啦.
2用到的包
rm(list = ls())
# devtools::install_github("myles-lewis/easylabel")
library(easylabel)
library(tidyverse)
library(qvalue)
library(AnnotationDbi)
library(org.Hs.eg.db)
library(ggstatsplot)
library(plotly)
3散点图
3.1 初步绘图
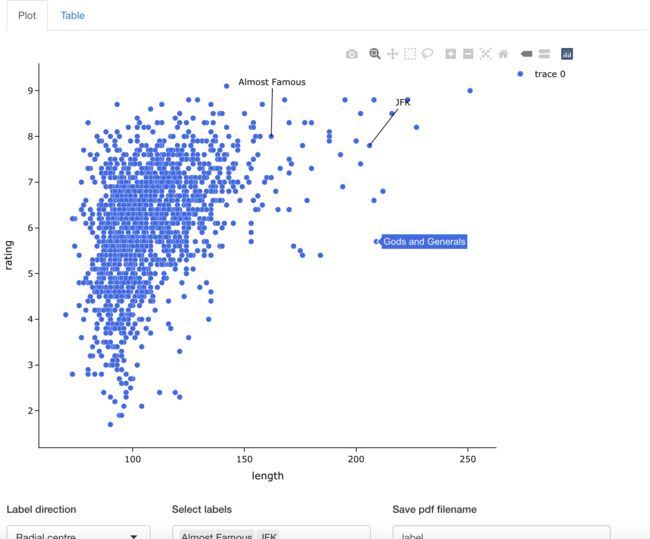
我们先简单画个散点图, 试着标记一下吧。
这里是可以交互的,大家直接点一下就标记了哦。
dat <- movies_long
dat %>%
easylabel(x = 'length',
y = 'rating',
labs = 'title',
colScheme = 'royalblue')
3.2 简单导出文件
接着在底部可以找到输出按钮,点击后导出你需要的格式吧~
3.3 导出为ploty对象
我们也可以通过设置output_shiny = F, 不激活shiny而直接导出为ploty对象, 依然是可交互的哦, 请随意移动。
p1 <- easylabel(dat,
x = 'length', y = 'rating', col = 'genre',
startLabels = dat$rating[dat$year == 1994],
output_shiny = F) %>%
layout(yaxis = list(zeroline = F))
p2 <- easylabel(dat,
x = 'length', y = 'votes', col = 'genre',
startLabels = dat$votes[dat$year == 1994],
output_shiny = F) %>%
layout(yaxis = list(zeroline = F))
plotly::subplot(p1, p2, nrows = 2, shareY = T, titleX = T, margin = 0.05)

4美化细节
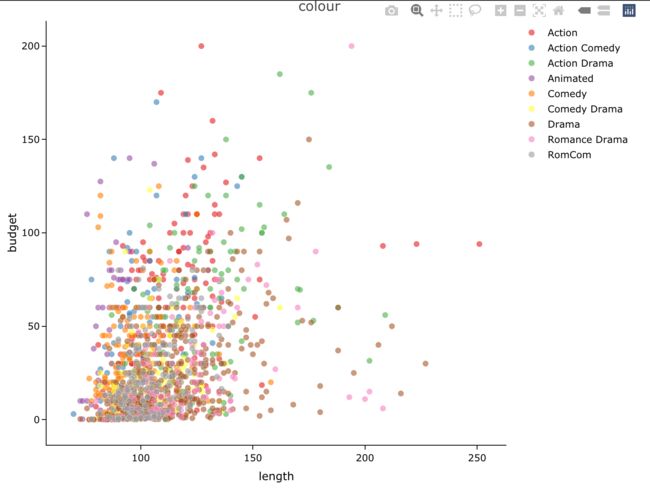
接着我们做一些细节的美化, 支持colour, shape, size等设置。
4.1 colour
easylabel(dat,
x = 'length', y = 'budget',
col = 'genre', alpha = 0.6,
output_shiny = F,
labs = 'title',
main = 'colour')%>%
layout(yaxis = list(zeroline = F))
4.2 shape
dat[1:100,] %>%
easylabel(
x = 'length', y = 'rating',
col = 'genre', alpha = 0.6,
shape = 'mpaa', shapeScheme = 21,
output_shiny = F,
labs = 'title',
main = 'shape') %>%
layout(yaxis = list(zeroline = F))

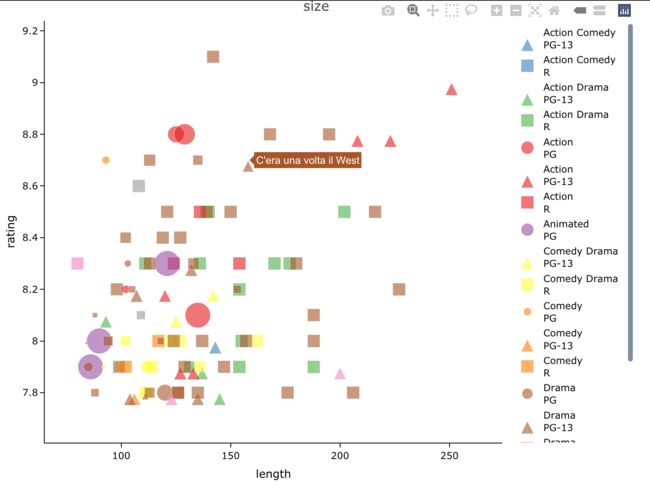
4.3 size
dat[1:100,] %>%
easylabel(
x = 'length', y = 'rating',
col = 'genre', alpha = 0.6,
shape = 'mpaa', shapeScheme = 21,
size = 'budget',
output_shiny = F,
labs = 'title',
main = 'size') %>%
layout(yaxis = list(zeroline = F))
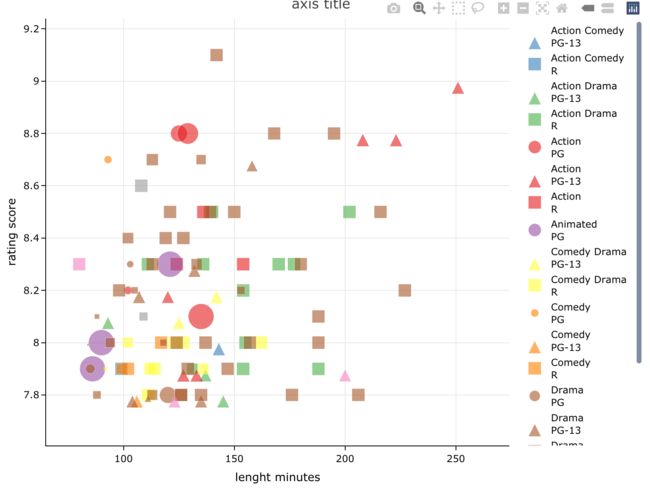
4.4 轴标题
dat[1:100,] %>%
easylabel(
x = 'length', y = 'rating',
col = 'genre', alpha = 0.6,
shape = 'mpaa', shapeScheme = 21,
size = 'budget',
output_shiny = F,
labs = 'title',
main = 'axis title',
xlab = 'lenght minutes',
ylab = 'rating score',
showgrid = T) %>%
layout(yaxis = list(zeroline = F))
5火山图
5.1 用到的数据
这里我们需要用到easyVolcano()函数, 输入文件为DESeq2, limma或EdgeR计算的差异基因结果。
这里我准备另一个我之前计算的limma结果。
all_diff <- read.csv("./alldiff_paired.csv",row.names = 1)
DT::datatable(all_diff)
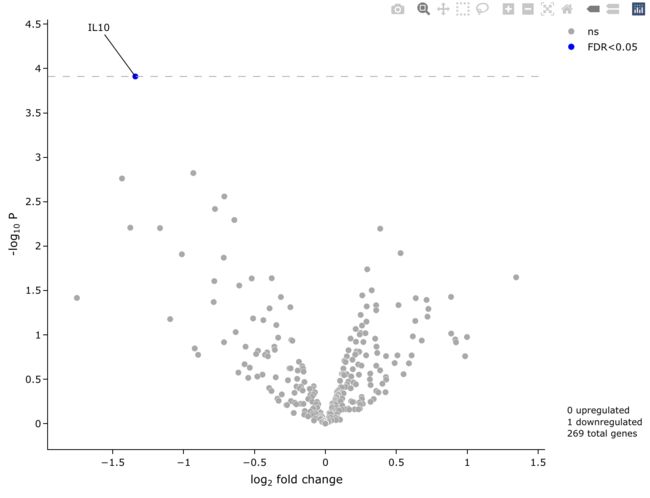
5.2 初步绘图
easyVolcano会使用自动识别DESeq2, limma或EdgeR计算的差异基因结果, 但默认是使用FDR。
easyVolcano(all_diff,useQ = T,
output_shiny = F,
startLabels = rownames(all_diff)[all_diff$adj.P.Val < 0.05])
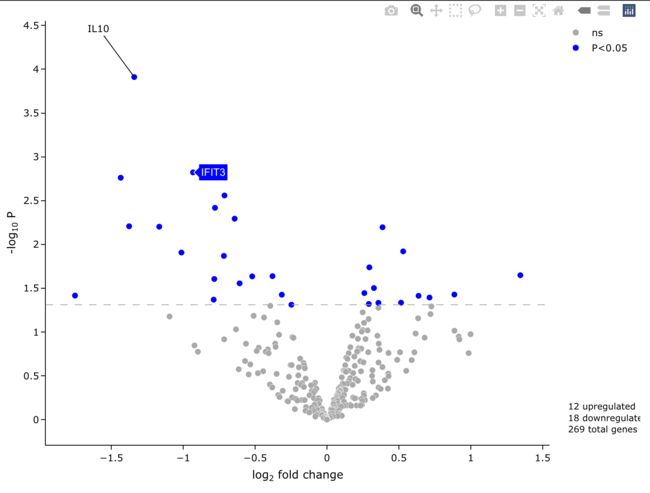
5.3 调整输入数据
手动选择x, y轴。
easyVolcano(all_diff,
useQ = F,
x = 'logFC',
y = 'P.Value',
startLabels = rownames(all_diff)[all_diff$adj.P.Val < 0.05],
output_shiny = F)
5.4 MA plot
需要用到easyMAplot函数, 也是非常容易上手.
easyMAplot(all_diff, useQ = T,
startLabels = rownames(all_diff)[all_diff$adj.P.Val < 0.05],
output_shiny = F)
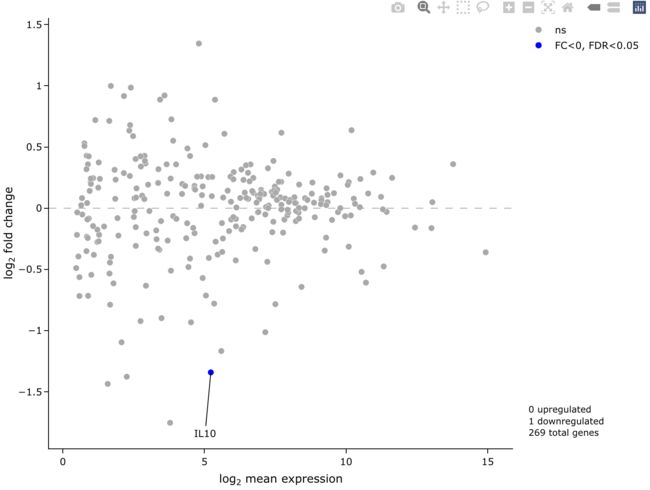
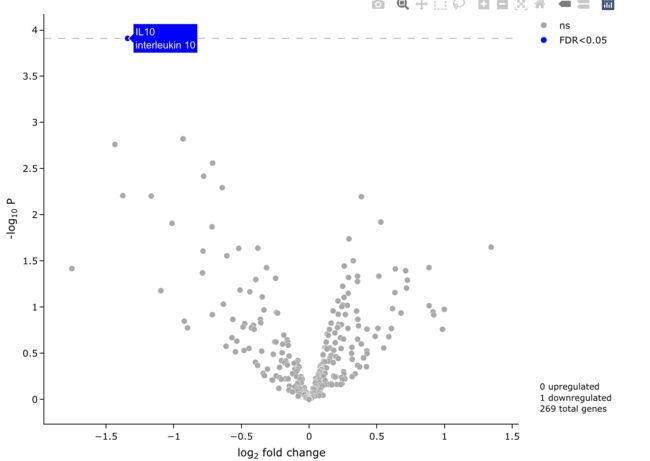
5.5 展示基因全名
有时候我们不光想展示Gene Symbol, 还想展示它的全名, 但是手动检索会非常麻烦, easyVolcano可以完美地帮你解决这个问题.
easyVolcano(all_diff, useQ = T,
fullGeneNames = T,
output_shiny = F
)
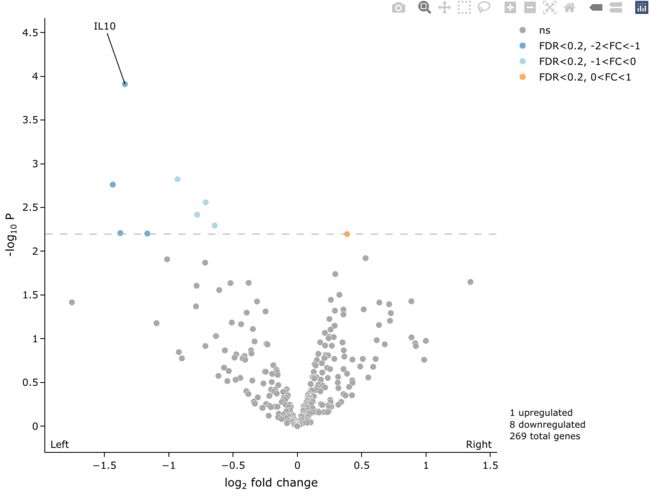
5.6 左右底角添加小标题
library(RColorBrewer)
colScheme <- c('darkgrey', brewer.pal(9, 'RdYlBu')[c(9:7, 3:1)])
easyVolcano(all_diff,
useQ = F, fullGeneNames = T,
Ltitle = expression(symbol("\254") ~ "Left"),
Rtitle = expression("Right" ~ symbol("\256")),
LRtitle_side = 1, # LRtitle_side = 1 bottom ; LRtitle_side = 3 top;
cex.lab = 0.9, cex.axis = 0.8,
fccut = c(1, 2), fdrcutoff = 0.2,
ylim = c(0, 6), xlim = c(-5, 5),
colScheme = colScheme,
startLabels = rownames(all_diff)[all_diff$adj.P.Val < 0.05],
output_shiny = F
)
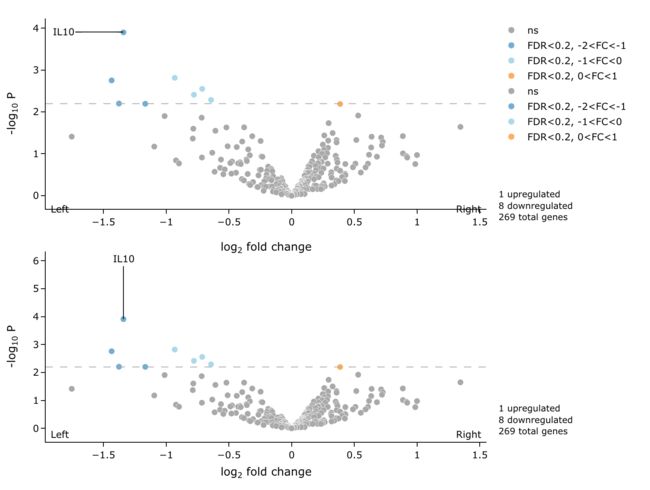
5.7 控制label方向
我们在这里示范一下水平方向和垂直方向.
p_horiz <- easyVolcano(all_diff,
useQ = F, fullGeneNames = T,
Ltitle = expression(symbol("\254") ~ "Left"),
Rtitle = expression("Right" ~ symbol("\256")),
LRtitle_side = 1, # LRtitle_side = 1 bottom ; LRtitle_side = 3 top;
cex.lab = 0.9, cex.axis = 0.8,
fccut = c(1, 2), fdrcutoff = 0.2,
ylim = c(0, 6), xlim = c(-5, 5),
colScheme = colScheme,
labelDir = "horiz",
startLabels = rownames(all_diff)[all_diff$adj.P.Val < 0.05],
output_shiny = F
)
p_vert <- easyVolcano(all_diff,
useQ = F, fullGeneNames = T,
Ltitle = expression(symbol("\254") ~ "Left"),
Rtitle = expression("Right" ~ symbol("\256")),
LRtitle_side = 1, # LRtitle_side = 1 bottom ; LRtitle_side = 3 top;
cex.lab = 0.9, cex.axis = 0.8,
fccut = c(1, 2), fdrcutoff = 0.2,
ylim = c(0, 6), xlim = c(-5, 5),
colScheme = colScheme,
labelDir = "vert",
startLabels = rownames(all_diff)[all_diff$adj.P.Val < 0.05],
output_shiny = F
)
plotly::subplot(p_horiz, p_vert, nrows = 2, shareY = T, titleX = T, margin = 0.05)
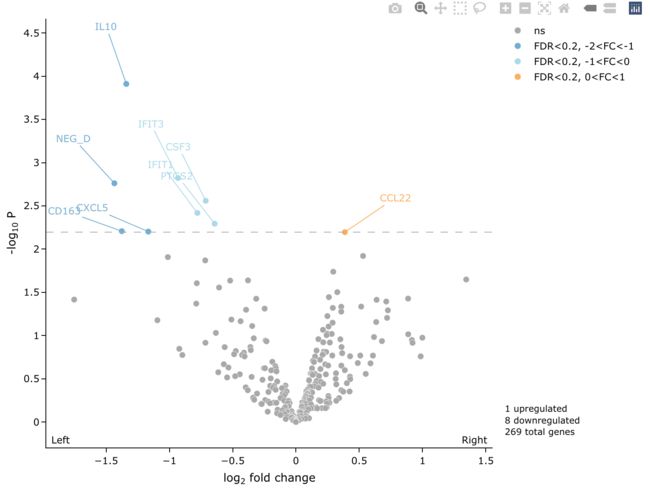
5.8 统一label和point的颜色
我们经常会遇到这种问题, 希望label和point统一颜色, 这样会更加美观。
这也提供了相应的解决办法, 设置text_col = "match"以及 line_col = "match"即可.
easyVolcano(all_diff,
useQ = F, fullGeneNames = T,
Ltitle = expression(symbol("\254") ~ "Left"),
Rtitle = expression("Right" ~ symbol("\256")),
LRtitle_side = 1, # LRtitle_side = 1 bottom ; LRtitle_side = 3 top;
cex.lab = 0.9, cex.axis = 0.8,
fccut = c(1, 2), fdrcutoff = 0.2,
ylim = c(0, 6), xlim = c(-5, 5),
colScheme = colScheme,
startLabels = rownames(all_diff)[all_diff$P.Value < 0.01],
line_col = "match", text_col = "match",
rectangles = T, border_col = NA,
rect_col = "match", border_radius = 20, padding = 5,
output_shiny = F
)
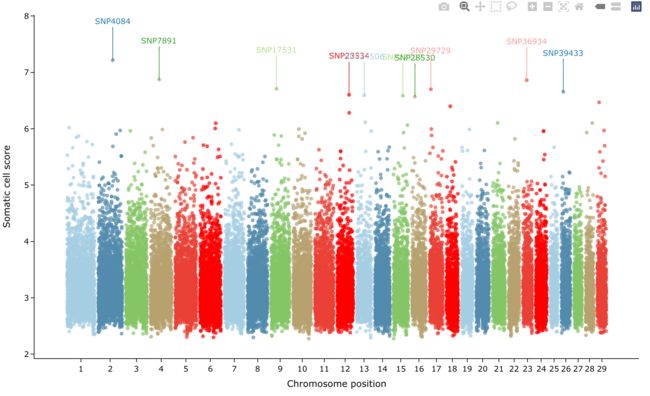
6曼哈顿图
这里我们也让point和label的颜色统一起来吧,颜值提升一下。
library(CMplot)
data("cattle50K")
chromCols <- RColorBrewer::brewer.pal(6, 'Paired')
easyManhattan(cattle50K,
chrom = "chr",
pos = "pos",
p = "Somatic cell score",
chromCols = chromCols,
ylab = "Somatic cell score",
output_shiny = F,
labs = "SNP",
npeaks = 10, ## 标注peak前10
labelDir = "vert",
line_col = "match", text_col = "match",
rectangles = T, border_col = NA,
rect_col = "match", border_radius = 20, padding = 5,
)

点个在看吧各位~ ✐.ɴɪᴄᴇ ᴅᴀʏ 〰
ComplexHeatmap | 颜狗写的高颜值热图代码!
ComplexHeatmap | 你的热图注释还挤在一起看不清吗!?
Google | 谷歌翻译崩了我们怎么办!?(附完美解决方案)
scRNA-seq | 吐血整理的单细胞入门教程
NetworkD3 | 让我们一起画个动态的桑基图吧~
RColorBrewer | 再多的配色也能轻松搞定!~
rms | 批量完成你的线性回归
CMplot | 完美复刻Nature上的曼哈顿图
Network | 高颜值动态网络可视化工具
boxjitter | 完美复刻Nature上的高颜值统计图
linkET | 完美解决ggcor安装失败方案(附教程)
......
本文由 mdnice 多平台发布