符号回归工具之 geppy: Python中的基因表达编程框架
符号回归工具之 geppy: Python中的基因表达编程框架
geppy是一个专门用于基因表达编程(GEP)的计算框架,由 C. Ferreira 在 2001 年提出 [1]。 geppy是在 Python 3 中开发的。这个框架个人认为稍微了解下遗传算法和遗传规划即可入门。
官网链接
什么是 GEP?
基因表达编程 (GEP) 是一种流行且成熟的进化算法,用于自动生成计算机程序和数学模型。它在符号回归、分类、自动模型设计、组合优化和实参数优化问题中得到了广泛的应用[2]。
基因表达式编程GEP(Gene Expression Programming)是一种基于生物基因结构和功能发明的一种新型自适应演化算法。
GEP可以看作是传统 遗传编程(GP)的变体,它使用固定长度的简单线性染色体来编码遗传信息。虽然染色体(基因)的长度是固定的,但由于其基因型-表型表达系统,它可以产生各种大小的表达树。许多实验表明,GEP 比 GP 更有效,并且由 GEP 进化的树的大小往往比 GP 的小。
geppy建立在优秀的进化计算框架DEAP之上,用于使用 GEP 进行快速原型设计和测试。DEAP 为 GP 提供基本支持,但缺乏对 GEP 的支持。geppy尽量遵循 DEAP 的风格,并试图保持与 DEAP 主要基础设施的兼容性。也就是说,geppy在某种程度上可以被认为是DEAP的一个插件,专门支持GEP。如果你熟悉 DEAP,那么很容易掌握geppy。此外,还提供全面的文档。
特征
- GEP中的核心数据结构,包括基因、染色体、表达树和K-表达。
- GEP中常见变异、转置、倒置和交叉算子的实现。
- 样板算法,包括标准 GEP 算法和集成局部优化器以进行数值常数优化的高级算法。
- 灵活的内置算法接口,可以支持任意数量的自定义变异和类交叉算子。
- 表达式树的可视化。
- 后处理中基因、染色体或 K 表达的符号简化。
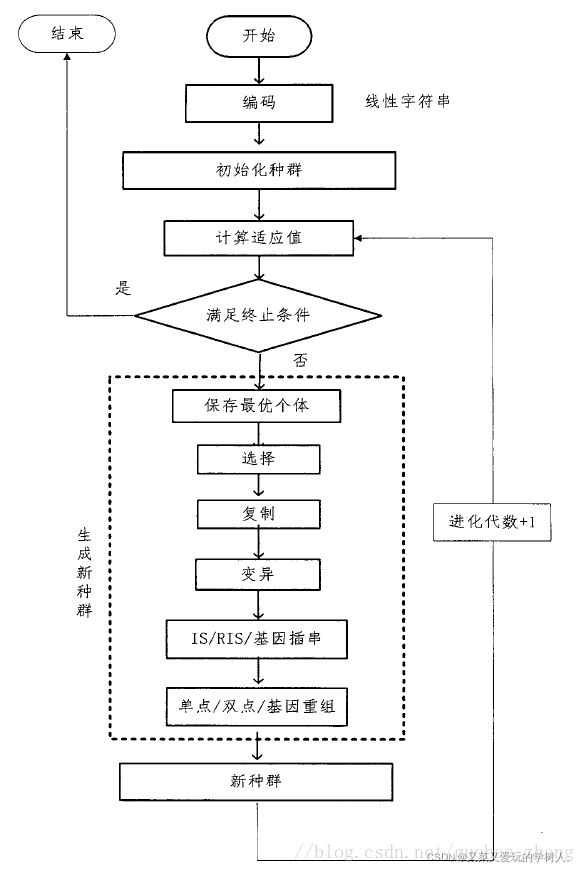
框架运行流程
- 数据集、观测数据的处理
- 创建基元集即定义输入变量的内容以及算子
- 注册基因、染色体和个体以及种群的基本参数
- 定义适应度评价函数
- 注册遗传算子
- 迭代进化(预测)
- 可视化
算法流程
-
基因 - 基础公式算子
-
个体(染色体) - 由多个基因组成即预测的公式
-
种群 - 所有预测的公式合集
-
变异 - 在交叉操作过后形成的新个体,有一定的概率会发生基因变异,与选择操作一样,这个操作是基于概率。
-
交叉 - 选择两个进行相互交配,将他们的染色体按照某种方式相互交换部分基因,形成两个新的个体的过程。
三个例子
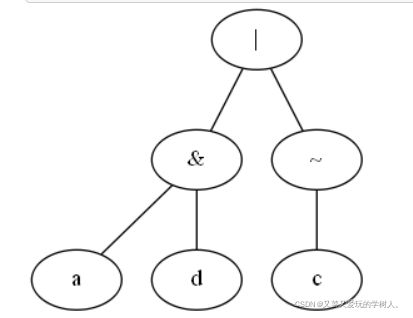
1.布尔函数的预测(与或非)
# 布尔值回归
# 定义数据集Y标签生成函数
def f(a, b, c, d):
""" The true model, which only involves three inputs on purpose."""
return (a and d) or not c
# 生成数据集[X,Y]
import itertools
X = []
Y = []
for a, b, c, d in itertools.product([True, False], repeat=4):
X.append((a, b, c, d))
Y.append(f(a, b, c, d))
# 创建geppy初始数据集
import geppy as gep
import operator
pset = gep.PrimitiveSet('Main', input_names=['a', 'b', 'c', 'd'])
# 加入算子
pset.add_function(operator.and_, 2)
pset.add_function(operator.or_, 2)
pset.add_function(operator.not_, 1)
from deap import creator, base, tools
creator.create("FitnessMax", base.Fitness, weights=(1,)) # 最大化目标函数
creator.create("Individual", gep.Chromosome, fitness=creator.FitnessMax)
h = 5 # 基因头长度
n_genes = 2 # 一条染色体中基因的长度
toolbox = gep.Toolbox()
toolbox.register('gene_gen', gep.Gene, pset=pset, head_length=h)
toolbox.register('individual', creator.Individual, gene_gen=toolbox.gene_gen, n_genes=n_genes, linker=operator.or_)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
# 将个人转化为可执行的lambda函数,即编译后可以得到预测值结果,再用于优化
toolbox.register('compile', gep.compile_, pset=pset)
def evaluate(individual):
"""Evalute the fitness of an individual"""
func = toolbox.compile(individual)
n_correct = 0
for (a, b, c, d), y in zip(X, Y):
prediction = func(a, b, c, d)
if prediction == y:
n_correct += 1
return n_correct,
toolbox.register('evaluate', evaluate)
toolbox.register('select', tools.selRoulette)
# 定义变异概率
toolbox.register('mut_uniform', gep.mutate_uniform, pset=pset, ind_pb=2 / (2 * h + 1))
toolbox.pbs['mut_uniform'] = 0.1
toolbox.register('mut_invert', gep.invert, pb=0.1)
toolbox.register('mut_is_ts', gep.is_transpose, pb=0.1)
toolbox.register('mut_ris_ts', gep.ris_transpose, pb=0.1)
toolbox.register('mut_gene_ts', gep.gene_transpose, pb=0.1)
# 定义交叉算子
toolbox.register('cx_1p', gep.crossover_one_point, pb=0.1)
toolbox.pbs['cx_1p'] = 0.4 # just show that the probability can be overwritten
toolbox.register('cx_2p', gep.crossover_two_point, pb=0.2)
toolbox.register('cx_gene', gep.crossover_gene, pb=0.1)
# 用于进化过程中评估函数中间结果的显示
import numpy
stats = tools.Statistics(key=lambda ind: ind.fitness.values[0])
stats.register("avg", numpy.mean)
stats.register("std", numpy.std)
stats.register("min", numpy.min)
stats.register("max", numpy.max)
import random
random.seed(123)
# 种群基本参数
n_pop = 50
n_gen = 50
pop = toolbox.population(n=n_pop)
hof = tools.HallOfFame(1) # 记录最好的个体
# 开始进化 - 预测
pop, log = gep.gep_simple(pop, toolbox,
n_generations=n_gen, n_elites=2,
stats=stats, hall_of_fame=hof, verbose=True)
best = hof[0]
symplified_best = gep.simplify(best)
print('\n')
print("符号回归结果:",symplified_best)
print('\n')
结果

最好的个人预测结果,可以看到其基因组成,基因是由geppy框架中toolbox注册,即一个基因由多少个算子组成。
- 个人认为最终的预测结果好坏可能需要去找到一些合适的算子和参数。
可视化
2.数值表达式的推理(只涉及整数常量,结果一般比较唯一)
基本流程与布尔函数预测一样,注册加减乘除算子,然后就是评估函数应该是最小化np.mean(np.abs(Y - Yp)) 与布尔函数不同。
import geppy as gep
from deap import creator, base, tools
import numpy as np
import random
s = 0
random.seed(s)
np.random.seed(s)
def f(x):
return -2 * x ** 2 + 11 * x + 1.8
n_cases = 100
X = np.random.uniform(-10, 10, size=n_cases)
Y = f(X) + np.random.normal(size=n_cases)
print(Y)
def protected_div(x1, x2):
if abs(x2) < 1e-6:
return 1
return x1 / x2
import operator
pset = gep.PrimitiveSet('Main', input_names=['x'])
pset.add_function(operator.add, 2)
pset.add_function(operator.sub, 2)
pset.add_function(operator.mul, 2)
pset.add_function(protected_div, 2)
pset.add_ephemeral_terminal(name='enc', gen=lambda: random.randint(-10, 10)) # each ENC is a random integer within [-10, 10]
from deap import creator, base, tools
creator.create("FitnessMin", base.Fitness, weights=(-1,)) # to minimize the objective (fitness)
creator.create("Individual", gep.Chromosome, fitness=creator.FitnessMin)
h = 7
n_genes = 2
toolbox = gep.Toolbox()
toolbox.register('gene_gen', gep.Gene, pset=pset, head_length=h)
toolbox.register('individual', creator.Individual, gene_gen=toolbox.gene_gen, n_genes=n_genes, linker=operator.add)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
toolbox.register('compile', gep.compile_, pset=pset)
def evaluate(individual):
func = toolbox.compile(individual)
Yp = np.array(list(map(func, X)))
return np.mean(np.abs(Y - Yp)),
toolbox.register('evaluate', evaluate)
toolbox.register('select', tools.selTournament, tournsize=3)
toolbox.register('mut_uniform', gep.mutate_uniform, pset=pset, ind_pb=0.05, pb=1)
toolbox.register('mut_invert', gep.invert, pb=0.1)
toolbox.register('mut_is_transpose', gep.is_transpose, pb=0.1)
toolbox.register('mut_ris_transpose', gep.ris_transpose, pb=0.1)
toolbox.register('mut_gene_transpose', gep.gene_transpose, pb=0.1)
toolbox.register('cx_1p', gep.crossover_one_point, pb=0.4)
toolbox.register('cx_2p', gep.crossover_two_point, pb=0.2)
toolbox.register('cx_gene', gep.crossover_gene, pb=0.1)
toolbox.register('mut_ephemeral', gep.mutate_uniform_ephemeral, ind_pb='1p')
toolbox.pbs['mut_ephemeral'] = 1
stats = tools.Statistics(key=lambda ind: ind.fitness.values[0])
stats.register("avg", np.mean)
stats.register("std", np.std)
stats.register("min", np.min)
stats.register("max", np.max)
n_pop = 100
n_gen = 100
pop = toolbox.population(n=n_pop)
hof = tools.HallOfFame(3)
# start evolution
pop, log = gep.gep_simple(pop, toolbox, n_generations=n_gen, n_elites=1,
stats=stats, hall_of_fame=hof, verbose=True)
best_ind = hof[0]
symplified_best = gep.simplify(best_ind)
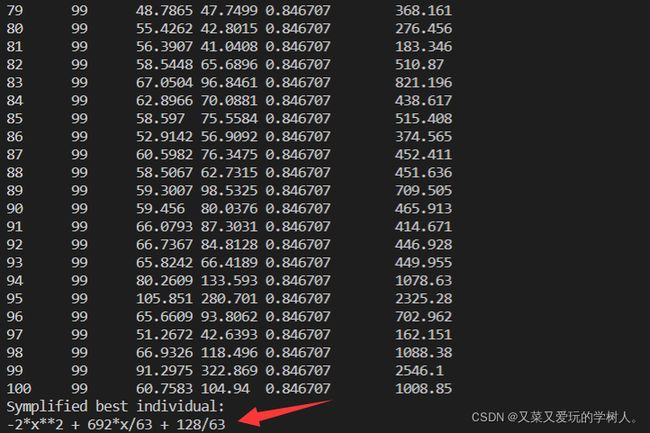
print('Symplified best individual: ')
print(symplified_best)
# 可视化
# import os
# os.environ["PATH"] += os.pathsep + 'D:/Program Files (x86)/Graphviz/bin/'
# rename_labels = {'add': '+', 'sub': '-', 'mul': '*', 'protected_div': '/'}
# gep.export_expression_tree(best_ind, rename_labels, 'data/numerical_expression_tree.png')
# from IPython.display import Image
# Image(filename='data/numerical_expression_tree.png')
结果
可视化
3.是用线性缩放的高阶符号回归(可以处理小数点型连续实数,结果可能不唯一)
import geppy as gep
from deap import creator, base, tools
import numpy as np
import random
s = 10
random.seed(s)
np.random.seed(s)
# 定义是否使用线性缩放
LINEAR_SCALING = True
def f(x):
return -2.45 * x ** 2 + 9.87 * x + 14.56
n_cases = 100
X = np.random.uniform(-10, 10, size=n_cases)
Y = f(X) + np.random.normal(size=n_cases)
Y = f(X)
def protected_div(a, b):
if np.isscalar(b):
if abs(b) < 1e-6:
b = 1
else:
b[abs(b) < 1e-6] = 1
return a / b
import operator
pset = gep.PrimitiveSet('Main', input_names=['x'])
pset.add_function(operator.add, 2)
pset.add_function(operator.sub, 2)
pset.add_function(operator.mul, 2)
pset.add_function(protected_div, 2)
pset.add_ephemeral_terminal(name='enc', gen=lambda: random.uniform(-5, 5))
from deap import creator, base, tools
creator.create("FitnessMin", base.Fitness, weights=(-1,)) # 最小化目标函数
creator.create("Individual", gep.Chromosome, fitness=creator.FitnessMin, a=float, b=float)
h = 7
n_genes = 2
toolbox = gep.Toolbox()
toolbox.register('gene_gen', gep.Gene, pset=pset, head_length=h)
toolbox.register('individual', creator.Individual, gene_gen=toolbox.gene_gen, n_genes=n_genes, linker=operator.add)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
toolbox.register('compile', gep.compile_, pset=pset)
def evaluate(individual):
func = toolbox.compile(individual)
Yp = func(X)
return np.mean((Y - Yp) ** 2), # 均方差
def evaluate_linear_scaling(individual):
func = toolbox.compile(individual)
Yp = func(X).
if isinstance(Yp, np.ndarray):
Q = np.hstack((np.reshape(Yp, (-1, 1)), np.ones((len(Yp), 1))))
(individual.a, individual.b), residuals, _, _ = np.linalg.lstsq(Q, Y, rcond=None)
if residuals.size > 0:
return residuals[0] / len(Y),
individual.a = 0
individual.b = np.mean(Y)
return np.mean((Y - individual.b) ** 2),
if LINEAR_SCALING:
toolbox.register('evaluate', evaluate_linear_scaling)
else:
toolbox.register('evaluate', evaluate)
toolbox.register('select', tools.selTournament, tournsize=3)
# 1. general operators
toolbox.register('mut_uniform', gep.mutate_uniform, pset=pset, ind_pb=0.05, pb=1)
toolbox.register('mut_invert', gep.invert, pb=0.1)
toolbox.register('mut_is_transpose', gep.is_transpose, pb=0.1)
toolbox.register('mut_ris_transpose', gep.ris_transpose, pb=0.1)
toolbox.register('mut_gene_transpose', gep.gene_transpose, pb=0.1)
toolbox.register('cx_1p', gep.crossover_one_point, pb=0.4)
toolbox.register('cx_2p', gep.crossover_two_point, pb=0.2)
toolbox.register('cx_gene', gep.crossover_gene, pb=0.1)
toolbox.register('mut_ephemeral', gep.mutate_uniform_ephemeral, ind_pb='1p')
toolbox.pbs['mut_ephemeral'] = 1
stats = tools.Statistics(key=lambda ind: ind.fitness.values[0])
stats.register("avg", np.mean)
stats.register("std", np.std)
stats.register("min", np.min)
stats.register("max", np.max)
n_pop = 100
n_gen = 200
pop = toolbox.population(n=n_pop)
hof = tools.HallOfFame(3)
pop, log = gep.gep_simple(pop, toolbox, n_generations=n_gen, n_elites=1,
stats=stats, hall_of_fame=hof, verbose=True)
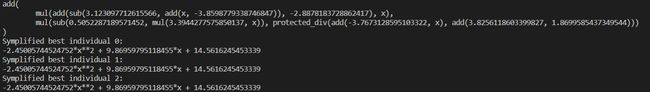
print(hof[0])
for i in range(3):
ind = hof[i]
symplified_model = gep.simplify(ind)
if LINEAR_SCALING:
symplified_model = ind.a * symplified_model + ind.b
print('Symplified best individual {}: '.format(i))
print(symplified_model)