机器学习——数据处理流程梳理
数据处理流程梳理
- 一、数据预处理
-
- 1.数据缺失处理
- 2.划分训练集、测试集及数据不平衡处理
- 3.数据标准化
- 4.特征选择
-
- 1)随机森林
- 2)相关性检测
- 3)lasso特征
- 5.特征降维
- 二、训练模型
-
- 1.训练
- 2.查看ROC曲线
- 3.评价指标
- 三、测试数据预处理
-
- 1.去除训练集缺失率高的列
- 2.标准化
- 3.特征选择
- 4.特征降维
- 四、测试集预测
- 五、完整代码
- 六、问题归纳
-
- 1.数据集划分、数据平衡处理的顺序
- 2.数据标准化应该放在哪一步
- 3.关键参数
-
- 1)行列缺失率row_miss_rate、col_miss_rate
- 2)模型参数
- 3)PCA保留的维度数
- 4.roc曲线随测试集样本比例而变化,各种指标表现过低
一、数据预处理
以二分类问题为例,数据集形式为excle,label在最后一列
1.数据缺失处理
分别对行列缺失进行处理
def preTrainDataAndSelectFeature(self, file_path='', label_name='', col_miss_rate=0.5,row_miss_rate=0.8,
balance_prior=False,all_data_balance=0.33,train_data_balance=0.33,test_data_balance=None,
forest_n=None, kbest_n=None, lasso=False, pca_feature=4, ):
"""
训练集数据处理
:param file_path:string 文件路径
:param label_name:标签列名
:param col_miss_rate:float 列缺失比率
:param row_miss_rate:float 标签为0的行缺失率
:param balance_prior:bool 数据集平衡优先
:param all_data_balance:balance_prior=True时,float 整体数据不均衡调整比例 若=1,为0,1标签比值为1:1,=0.33为1:0.33
:param train_data_balance:balance_prior=False时,
:param test_data_balance:balance_prior=False时,
:param forest_n:int 使用forest选取的特征数
:param kbest_n:int 使用kbest选取的特征数
:param lasso:bool 是否使用lasso回归选取特征
:param pca_feature:int pca降维后的特征数(这是降维的最后一步,数据此后一直使用降维后的特征)
:return:可直接放入模型训练的最终数据
"""
data = pd.read_excel(file_path)
# 列处理
data = data.dropna(axis=1, thresh=data.shape[0] * col_miss_rate)
# 保存过滤缺失后的特征
self.drop_select = list(data)
self.final_feature = np.array(self.drop_select)
print("drop select:",len(self.final_feature),"\n", self.final_feature)
# 行处理:根据标签取值拆分样本,不同取值用各自的均值填充
data_group = data.groupby(label_name)
data0 = dict([x for x in data_group])[0]
data0 = data0.dropna(axis=0, thresh=data0.shape[1] * row_miss_rate)
data0 = data0.fillna(data0.mean())
data1 = dict([x for x in data_group])[1]
data1 = data1.fillna(data1.mean())
data = pd.concat([data0, data1], axis=0)
2.划分训练集、测试集及数据不平衡处理
这里存在两个问题:
1.是否用整体的数据集先提取特征,再应用特征,然后划分训练集测试集?
这样可以较好的考虑到数据集的整体特征,但同时,测试集的表现可能会高于真实场景,考虑到数据集较大,故没有使用该方法。
2.数据平衡处理、数据集划分顺序
两种做法:先整体数据集平衡处理,再划分训练集、测试集 或 先划分训练集、测试集,再单独对训练集进行平衡处理(,若测试集样本比例极不平衡,也需要平衡处理)。
同样,由于第一种方法测试集表现可能过优,采用后一种。
这里,将两种方法进行实现,balance_prior=True,为第一种,否则第二种。
# # 数据不平衡处理
X, Y = data.drop([label_name], axis=1), data[label_name]
if balance_prior:
#先数据平衡处理,再拆分训练集、测试集
smo = BorderlineSMOTE(kind='borderline-1',
sampling_strategy={0: Y.value_counts()[0], 1: int(Y.value_counts()[0] * all_data_balance)},
random_state=2)
x, y = smo.fit_resample(X, Y)
print("data balance result:\n", pd.Series(y).value_counts())
# 打乱不平衡后的数据
data_select = pd.concat((x, y), axis=1)
data_select = shuffle(data_select,random_state=1)
X, Y = data_select.drop([label_name], axis=1), data_select[label_name]
#划分训练集、测试集
X_train, X_test, y_train, y_test = model_selection.train_test_split(X,Y, test_size=0.3, random_state=1234)
else:
# 先拆分训练集、测试集,再分别对训练集、测试集平衡处理
X_train, X_test, y_train, y_test = model_selection.train_test_split(X, Y, test_size=0.3, random_state=1234)
# # 数据不平衡处理
smo = BorderlineSMOTE(kind='borderline-1',
sampling_strategy={0: y_train.value_counts()[0],
1: int(y_train.value_counts()[0] * train_data_balance)},
random_state=2)
x_train, y_train = smo.fit_resample(X_train, y_train)
print("train data balance result:\n", pd.Series(y_train).value_counts())
if test_data_balance is not None:
smo = BorderlineSMOTE(kind='borderline-1',
sampling_strategy={0: y_test.value_counts()[0],
1: int(y_test.value_counts()[0] * test_data_balance)},
random_state=2)
X_test, y_test = smo.fit_resample(X_test, y_test)
print("test data balance result:\n", pd.Series(y_test).value_counts())
# # 打乱平衡处理后的数据
data_select = pd.concat((x_train, y_train), axis=1)
data_select = shuffle(data_select, random_state=1)
X_train, y_train = data_select.drop([label_name], axis=1), data_select[label_name]
print("data train:\n", pd.Series(y_train).value_counts())
print("data test:\n", pd.Series(y_test).value_counts())
3.数据标准化
# 用训练集的数据特征对测试集标准化
x_train= pd.DataFrame(self.coder.fit_transform(X_train.values))
x_test =pd.DataFrame(self.coder.transform(X_test.values))
4.特征选择
在样本极其不平衡时,先将数据平衡处理再进行特征选择,选择的特征比较稳定。
1)随机森林
# 随机森林
if forest_n!=None:
forest = RandomForestClassifier(random_state=1)
forest.fit(x_train, y_train)
ra=pd.Series(forest.feature_importances_,index=x_train.columns)
self.forest_select= ra.sort_values()[::-1][:forest_n].index
print("forest importance:",forest.feature_importances_)
x_train = x_train.loc[:, self.forest_select]
x_test = x_test.loc[:, self.forest_select]
self.final_feature=self.final_feature[self.forest_select]
print("forest features:", len(self.final_feature), "\n", self.final_feature)
2)相关性检测
# 相关性检测
if kbest_n != None:
pearsonr_way = lambda x, y: np.array(list(map(lambda x: pearsonr(np.squeeze(x), np.squeeze(y)), x.T))).T[0]
self.pearsonr_select = SelectKBest(pearsonr_way, k=kbest_n).fit(x_train, y_train).get_support(indices=True)
x_train = x_train.loc[:, self.pearsonr_select]
x_test = x_test.loc[:, self.pearsonr_select]
self.final_feature = self.final_feature[self.pearsonr_select]
print("kbest feature:",len(self.final_feature ),"\n", self.final_feature )
3)lasso特征
# lasso特征
if lasso:
# 交叉验证找出最好的lambdas
Lambdas = np.logspace(-5, 2, 200)
lasso_cv = LassoCV(alphas=Lambdas, normalize=True, cv=10, max_iter=10000)
lasso_cv.fit(x_train,y_train)
lasso = Lasso(alpha=lasso_cv.alpha_, normalize=True, max_iter=10000)
lasso.fit(x_train,y_train)
print("lasso importance:\n",lasso.coef_,"\n","lasso mse:",mean_squared_error(y_train, lasso.predict(x_train)))
tmp = pd.Series(index=x_train.columns.tolist(), data=lasso.coef_.tolist())
self.lasso_select = x_train.columns[tmp.values != 0]
x_train = x_train.loc[:, self.lasso_select]
x_test= x_test.loc[:,self.lasso_select]
self.final_feature = self.final_feature[self.lasso_select]
print("lasso select:", len(self.final_feature), "\n", self.final_feature)
5.特征降维
使用PCA主成分分析
# 主成分分析降维
if pca_feature!=None:
self.pca = PCA(n_components=pca_feature)
self.pca.fit(x_train)
print("pca importance:", self.pca.explained_variance_ratio_)
x_train = self.pca.transform(x_train)
x_test=self.pca.transform(x_test)
return x_train, y_train,x_test,y_test
二、训练模型
1.训练
def Train(self, file_path='', label_name="", col_miss_rate=0.5, row_miss_rate=0.8,
balance_prior=False, all_data_balance=0.33, train_data_balance=1, test_data_balance=None,
forest_n=20, kbest_n=None, lasso=False, pca_feature=None, ):
x_train, y_train,x_test,y_test = self.preTrainDataAndSelectFeature(file_path=file_path, label_name=label_name,
col_miss_rate=col_miss_rate,row_miss_rate=row_miss_rate,
balance_prior=balance_prior, all_data_balance=all_data_balance,train_data_balance=train_data_balance, test_data_balance=test_data_balance,
forest_n=forest_n,kbest_n=kbest_n,lasso=lasso, pca_feature=pca_feature)
self.svm.fit(x_train, y_train)
2.查看ROC曲线
用predict_proba()进行预测,得到各类别概率值
self._showROC(self.svm.predict_proba(x_test)[:, 1], y_test)
def _showROC(self, y_pred, y_true):
fpr, tpr, thresholds = roc_curve(y_true,y_pred)
roc_auc = auc(fpr, tpr)
print("AUC:",roc_auc)
plt.plot(fpr, tpr, 'b', label='Val AUC = %0.3f' % roc_auc)
plt.legend(loc='upper right')
plt.ylabel('True Positive Rate')
plt.xlabel('False Positive Rate')
plt.show()
3.评价指标
y_pre=self.svm.predict(x_test)
print("recall_score:", recall_score(y_test, y_pre))
print("precision_score:", precision_score(y_test, y_pre))
print("f1_score:", f1_score(y_test, y_pre))
三、测试数据预处理
1.去除训练集缺失率高的列
根据训练集的处理步骤,使其在标准化前拥有相同的特征
def preEvalData(self,file_path="",label_name=""):
x= pd.read_excel(file_path)
#填充,若填充后仍有NAN,对x.mean()进行填充.fillna(0)
x = x.fillna(x.mean().fillna(0))
#去缺失特征和标签
x=x.loc[:, self.drop_select]
x = x.drop([label_name], axis=1)
2.标准化
# 用训练集的均值和方差进行标准化
x= pd.DataFrame(self.coder.transform(x.values))
3.特征选择
if self.forest_select is not None:
x = x.loc[:, self.forest_select]
if self.lasso_select is not None:
x = x.loc[:, self.lasso_select]
if self.pearsonr_select is not None:
x = x.loc[:, self.pearsonr_select]
4.特征降维
if self.pca is not None:
x = self.pca.transform(x.values)
return x
四、测试集预测
def predict(self, file_path='', label_name=""):
x=self.preEvalData(file_path,label_name=label_name)
pred=self.svm.predict_proba(x)
count_1 = 0
for i in pred[:,1]:
#观察roc曲线得阈值为0.8
if i >0.8 :
count_1 = count_1 + 1
print("1:", count_1)
print("0:", len(pred) - count_1)
五、完整代码
# coding=utf-8
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from imblearn.over_sampling import BorderlineSMOTE
from sklearn.preprocessing import StandardScaler
from sklearn.utils import shuffle
from sklearn.feature_selection import SelectKBest
from sklearn.linear_model import LassoCV,Lasso
from sklearn.ensemble import RandomForestClassifier
from sklearn.decomposition import PCA
from sklearn import svm,model_selection
from sklearn.metrics import mean_squared_error,roc_curve, auc,recall_score, precision_score, f1_score, roc_curve,accuracy_score
from scipy.stats import pearsonr
class ML():
def __init__(self):
#特征、数据处理参数,用preTrainDataAndSelectFeature()计算出实际值
#数据标准化模型,将训练集的均值方差应用到测试集
self.coder=StandardScaler()
#依据列缺失特征选择
self.drop_select=None
#随机森林特征选择
self.forest_select=None
#lasso回归特征选择
self.lasso_select=None
#kbest相关性检测特征选择
self.pearsonr_select=None
#pca数据降维模型
self.pca= None
# svc支持向量机模型
self.svm = svm.SVC(C=0.8, kernel='rbf', gamma=20, decision_function_shape='ovr', probability=True)
#合并特征,为中文信息、非索引
self.final_feature=None
def preTrainDataAndSelectFeature(self, file_path="", label_name='', col_miss_rate=0.5,row_miss_rate=0.8,
balance_prior=False,all_data_balance=0.33,train_data_balance=0.33,test_data_balance=None,
forest_n=None, kbest_n=None, lasso=False, pca_feature=4, ):
"""
训练集数据处理
:param file_path:string 文件路径
:param label_name:标签列名
:param col_miss_rate:float 列缺失比率
:param row_miss_rate:float 标签为0的行缺失率
:param balance_prior:bool 数据集平衡优先
:param all_data_balance:balance_prior=True时,float 整体数据不均衡调整比例 若=1,为0,1标签比值为1:1,=0.33为1:0.33
:param train_data_balance:balance_prior=False时,
:param test_data_balance:balance_prior=False时,
:param forest_n:int 使用forest选取的特征数
:param kbest_n:int 使用kbest选取的特征数
:param lasso:bool 是否使用lasso回归选取特征
:param pca_feature:int pca降维后的特征数(这是降维的最后一步,数据此后一直使用降维后的特征)
:return:可直接放入模型训练的最终数据
"""
data = pd.read_excel(file_path)
# 对数据缺失进行处理
data = data.dropna(axis=1, thresh=data.shape[0] * col_miss_rate)
# 保存过滤缺失后的特征
self.drop_select = list(data)
self.final_feature = np.array(self.drop_select)
print("drop select:",len(self.final_feature),"\n", self.final_feature)
# 行处理:根据标签取值拆分样本,不同取值用各自的均值填充
data_group = data.groupby(label_name)
data0 = dict([x for x in data_group])[0]
data0 = data0.dropna(axis=0, thresh=data0.shape[1] * row_miss_rate)
data0 = data0.fillna(data0.mean())
data1 = dict([x for x in data_group])[1]
data1 = data1.fillna(data1.mean())
data = pd.concat([data0, data1], axis=0)
# # 数据不平衡处理
X, Y = data.drop([label_name], axis=1), data[label_name]
if balance_prior:
#先数据平衡处理,再拆分训练集、测试集
smo = BorderlineSMOTE(kind='borderline-1',
sampling_strategy={0: Y.value_counts()[0], 1: int(Y.value_counts()[0] * all_data_balance)},
random_state=2)
x, y = smo.fit_resample(X, Y)
print("data balance result:\n", pd.Series(y).value_counts())
# 打乱不平衡后的数据
data_select = pd.concat((x, y), axis=1)
data_select = shuffle(data_select,random_state=1)
X, Y = data_select.drop([label_name], axis=1), data_select[label_name]
#划分训练集、测试集
X_train, X_test, y_train, y_test = model_selection.train_test_split(X,Y, test_size=0.3, random_state=1234)
else:
# 先拆分训练集、测试集,再分别对训练集、测试集平衡处理
X_train, X_test, y_train, y_test = model_selection.train_test_split(X, Y, test_size=0.3, random_state=1234)
# # 数据不平衡处理
smo = BorderlineSMOTE(kind='borderline-1',
sampling_strategy={0: y_train.value_counts()[0],
1: int(y_train.value_counts()[0] * train_data_balance)},
random_state=2)
x_train, y_train = smo.fit_resample(X_train, y_train)
print("train data balance result:\n", pd.Series(y_train).value_counts())
if test_data_balance is not None:
smo = BorderlineSMOTE(kind='borderline-1',
sampling_strategy={0: y_test.value_counts()[0],
1: int(y_test.value_counts()[0] * test_data_balance)},
random_state=2)
X_test, y_test = smo.fit_resample(X_test, y_test)
print("test data balance result:\n", pd.Series(y_test).value_counts())
# # 打乱平衡处理后的数据
data_select = pd.concat((x_train, y_train), axis=1)
data_select = shuffle(data_select, random_state=1)
X_train, y_train = data_select.drop([label_name], axis=1), data_select[label_name]
print("data train:\n", pd.Series(y_train).value_counts())
print("data test:\n", pd.Series(y_test).value_counts())
# 用训练集的数据特征对测试集标准化
x_train= pd.DataFrame(self.coder.fit_transform(X_train.values))
x_test =pd.DataFrame(self.coder.transform(X_test.values))
# 随机森林
if forest_n!=None:
forest = RandomForestClassifier(random_state=1)
forest.fit(x_train, y_train)
ra=pd.Series(forest.feature_importances_,index=x_train.columns)
self.forest_select= ra.sort_values()[::-1][:forest_n].index
print("forest importance:",forest.feature_importances_)
x_train = x_train.loc[:, self.forest_select]
x_test = x_test.loc[:, self.forest_select]
self.final_feature=self.final_feature[self.forest_select]
print("forest features:", len(self.final_feature), "\n", self.final_feature)
# 相关性检测
if kbest_n != None:
pearsonr_way = lambda x, y: np.array(list(map(lambda x: pearsonr(np.squeeze(x), np.squeeze(y)), x.T))).T[0]
self.pearsonr_select = SelectKBest(pearsonr_way, k=kbest_n).fit(x_train, y_train).get_support(indices=True)
x_train = x_train.loc[:, self.pearsonr_select]
x_test = x_test.loc[:, self.pearsonr_select]
self.final_feature = self.final_feature[self.pearsonr_select]
print("kbest feature:",len(self.final_feature ),"\n", self.final_feature )
# lasso特征
if lasso:
# 交叉验证找出最好的lambdas
Lambdas = np.logspace(-5, 2, 200)
lasso_cv = LassoCV(alphas=Lambdas, normalize=True, cv=10, max_iter=10000)
lasso_cv.fit(x_train,y_train)
lasso = Lasso(alpha=lasso_cv.alpha_, normalize=True, max_iter=10000)
lasso.fit(x_train,y_train)
print("lasso importance:\n",lasso.coef_,"\n","lasso mse:",mean_squared_error(y_train, lasso.predict(x_train)))
tmp = pd.Series(index=x_train.columns.tolist(), data=lasso.coef_.tolist())
self.lasso_select = x_train.columns[tmp.values != 0]
x_train = x_train.loc[:, self.lasso_select]
x_test= x_test.loc[:,self.lasso_select]
self.final_feature = self.final_feature[self.lasso_select]
print("lasso select:", len(self.final_feature), "\n", self.final_feature)
# 主成分分析降维
if pca_feature!=None:
self.pca = PCA(n_components=pca_feature)
self.pca.fit(x_train)
print("pca importance:", self.pca.explained_variance_ratio_)
x_train = self.pca.transform(x_train)
x_test=self.pca.transform(x_test)
return x_train, y_train,x_test,y_test
def preEvalData(self,file_path="",label_name=""):
x = pd.read_excel(file_path)
#填充,若填充后仍有NAN,对x.mean()进行填充.fillna(0)
x = x.fillna(x.mean().fillna(0))
#去缺失特征和标签
x=x.loc[:, self.drop_select]
x = x.drop([label_name], axis=1)
# 用训练集的均值和方差进行标准化
x= pd.DataFrame(self.coder.transform(x.values))
if self.forest_select is not None:
x = x.loc[:, self.forest_select]
if self.lasso_select is not None:
x = x.loc[:, self.lasso_select]
if self.pearsonr_select is not None:
x = x.loc[:, self.pearsonr_select]
if self.pca is not None:
x = self.pca.transform(x.values)
return x
return x.values
def Train(self, file_path='', label_name="", col_miss_rate=0.5, row_miss_rate=0.8,
balance_prior=False, all_data_balance=0.33, train_data_balance=1, test_data_balance=None,
forest_n=20, kbest_n=None, lasso=False, pca_feature=None, ):
x_train, y_train,x_test,y_test = self.preTrainDataAndSelectFeature(file_path=file_path, label_name=label_name,
col_miss_rate=col_miss_rate,row_miss_rate=row_miss_rate,
balance_prior=balance_prior, all_data_balance=all_data_balance,train_data_balance=train_data_balance, test_data_balance=test_data_balance,
forest_n=forest_n,kbest_n=kbest_n,lasso=lasso, pca_feature=pca_feature)
self.svm.fit(x_train, y_train)
self._showROC(self.svm.predict_proba(x_test)[:, 1], y_test)
y_pre=self.svm.predict(x_test)
print("recall_score:", recall_score(y_test, y_pre))
print("precision_score:", precision_score(y_test, y_pre))
print("f1_score:", f1_score(y_test, y_pre))
def predict(self, file_path='', label_name=""):
x=self.preEvalData(file_path,label_name=label_name)
pred=self.svm.predict_proba(x)
count_1 = 0
for i in pred[:,1]:
#观察roc曲线得阈值为0.8
if i >0.8 :
count_1 = count_1 + 1
print("1:", count_1)
print("0:", len(pred) - count_1)
def _showROC(self, y_pred, y_true):
fpr, tpr, thresholds = roc_curve(y_true,y_pred)
roc_auc = auc(fpr, tpr)
print("AUC:",roc_auc)
plt.plot(fpr, tpr, 'b', label='Val AUC = %0.3f' % roc_auc)
plt.legend(loc='upper right')
plt.ylabel('True Positive Rate')
plt.xlabel('False Positive Rate')
plt.show()
if __name__=="__main__":
pred=ML()
pred.Train(file_path='.\xxxxxx.xlsx', label_name='是否真实', col_miss_rate=0.5, row_miss_rate=0.8,
balance_prior=False, all_data_balance=None, train_data_balance=0.33, test_data_balance=0.1,
forest_n=None, kbest_n=None, lasso=True, pca_feature=6, )
#用训练好的模型预测
pred.predict(file_path=r'xxxx.xlsx', label_name='是否真实')
六、问题归纳
1.数据集划分、数据平衡处理的顺序
2.数据标准化应该放在哪一步
3.关键参数
示例参数
col_miss_rate=0.2, row_miss_rate=0.65,
balance_prior=False, all_data_balance=None, train_data_balance=0.33, test_data_balance=None,
forest_n=None, kbest_n=None, lasso=True, pca_feature=5,
1)行列缺失率row_miss_rate、col_miss_rate
较大可能错过关键特征,较小可能包含没有价值的数据,对结果影响很大
2)模型参数
C,gamma等
3)PCA保留的维度数
pca_feature
4.roc曲线随测试集样本比例而变化,各种指标表现过低
1)检查每次运行筛选的特征是否相同,降维的模型参数是否相同,这是ROC变化过大的主要原因,很多模型具有随机性,每次运行可能会选出不同的特征,可设置random_state属性解决。
2)测试集数据样本极其不平衡(但不到万不得已,最好不改变测试集)
如下,样本比例小于10:1时,roc变化较小,指标稳定
当测试集标签比例0:1约为130:1时:
ROC曲线:
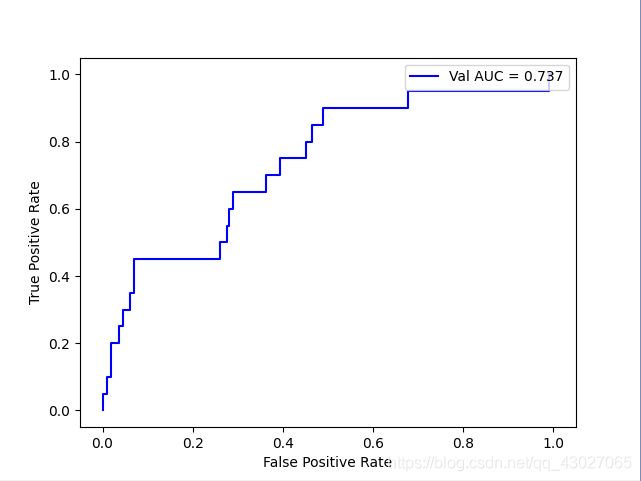
AUC: 0.7369069343065693
recall_score: 0.2
precision_score: 0.04938271604938271
f1_score: 0.07920792079207921
50:1
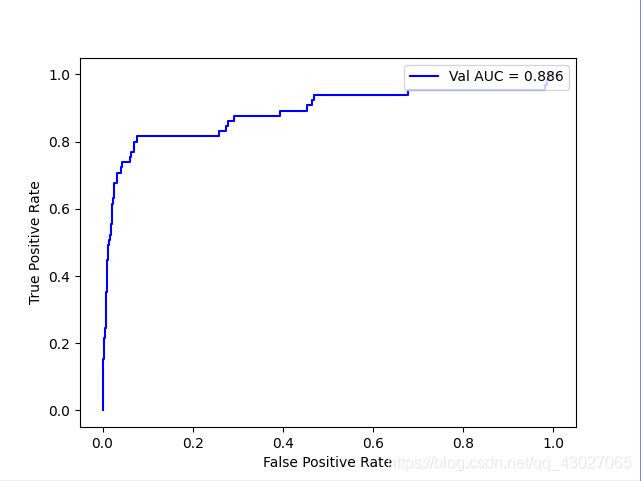
AUC: 0.8855932996443946
recall_score: 0.6153846153846154
precision_score: 0.3448275862068966
f1_score: 0.44198895027624313
10:1
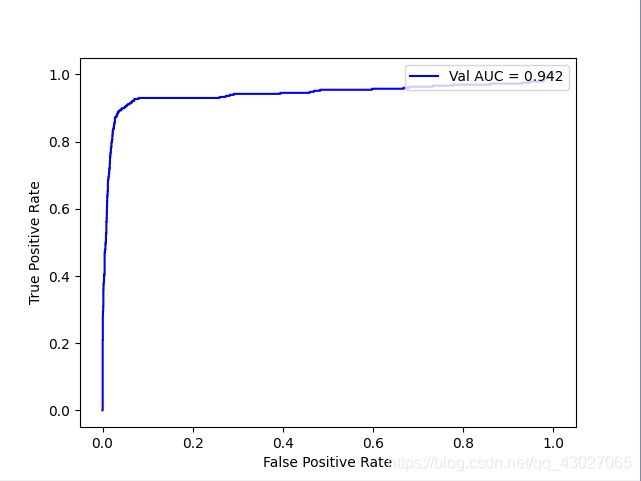
AUC: 0.9421023789389352
recall_score: 0.8353658536585366
precision_score: 0.7806267806267806
f1_score: 0.8070692194403534
3:1
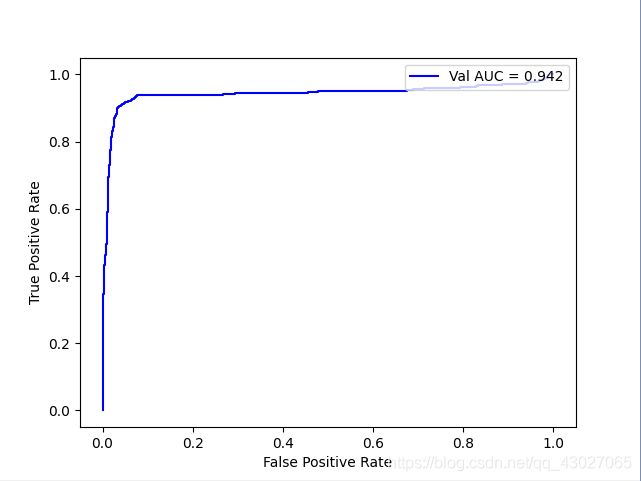
AUC: 0.9422224401968188
recall_score: 0.8407707910750507
precision_score: 0.9129955947136564
f1_score: 0.8753959873284055