肾病分析预测(机器学习_K最近邻分类器)
数据集链接:Chronic KIdney Disease dataset | Kaggle
导入python库
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import GridSearchCV, train_test_split, cross_val_score
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import r2_score
import scipy.stats as stats
import seaborn as sns
import missingno as msno
%matplotlib inline加载数据
df = pd.read_csv('kidney_disease.csv')
df.drop('id',axis=1,inplace=True)
df['classification'].replace(to_replace="ckd\t" ,value="ckd",inplace=True)
data = df
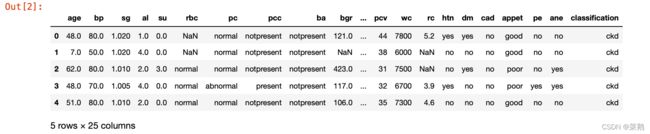
data.head()df.shape数据集预处理
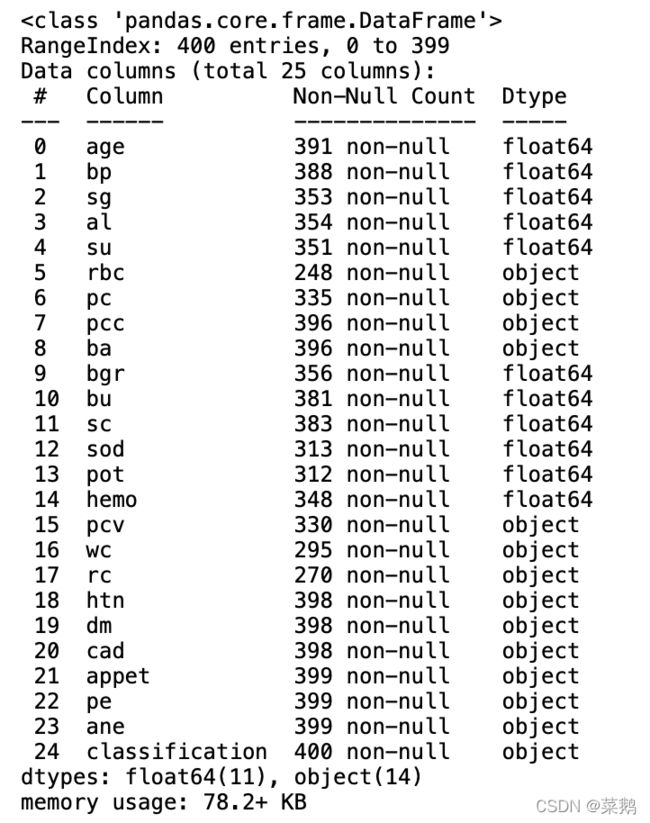
df.info()统计数据
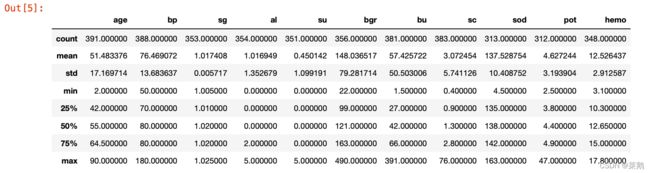
df.describe()
缺失值统计
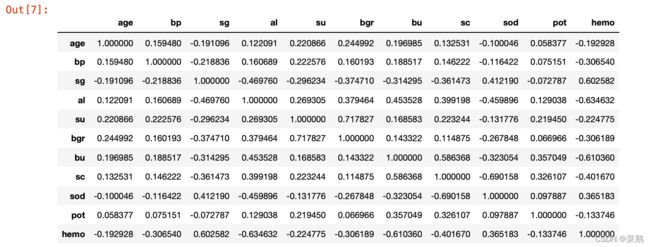
df.isna().sum()相关矩阵和矩阵可视化
df.corr()查看classification属性类型个数
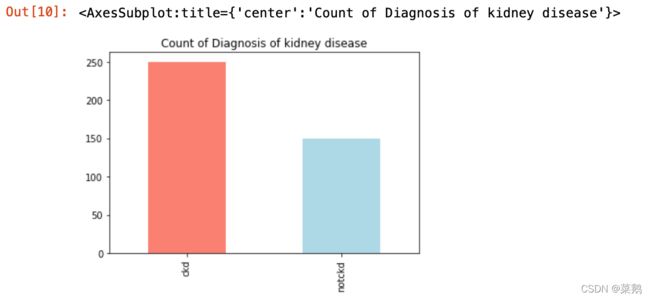
df['classification'].value_counts()推断: 从以上的输出中,可以推断出数据接近于“不平衡数据集”
countNoDisease = len(df[df['classification'] == 0])
countHaveDisease = len(df[df['classification'] == 1])
print("Percentage of Patients Haven't Heart Disease: {:.2f}%".format(
(countNoDisease / (len(df['classification']))*100)))
print("Percentage of Patients Have Heart Disease: {:.2f}%".format(
(countHaveDisease / (len(df['classification']))*100)))通过图表分析数据的平衡性
df['classification'].value_counts().plot(kind='bar', color=['salmon', 'lightblue'],
title="Count of Diagnosis of kidney disease")分析“年龄”列的分布
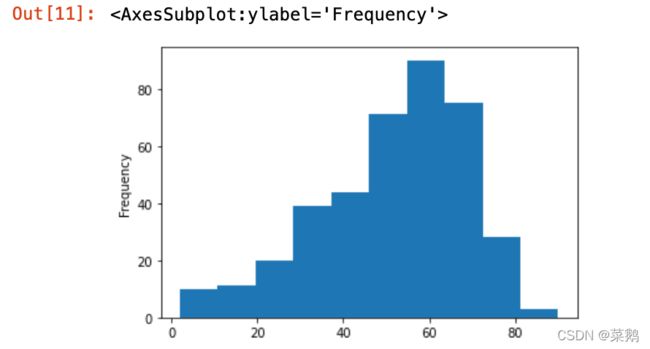
df['age'].plot(kind='hist')推论: 根据直方图,可推断50-60岁年龄段的人数更多。
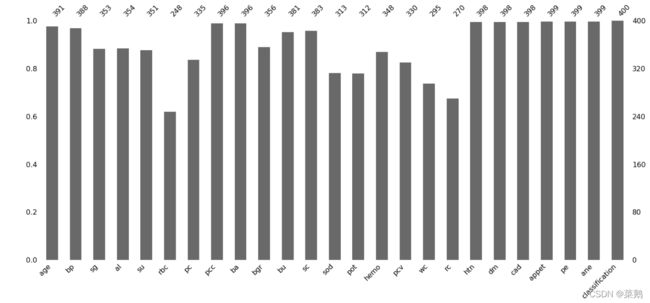
p = msno.bar(df)推论: 这里任何未触及顶部 400 标记的特征都具有空值。
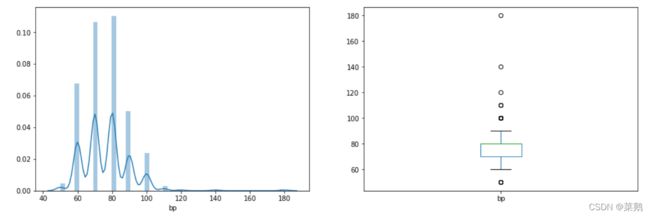
plt.subplot(121), sns.distplot(df['bp'])
plt.subplot(122), df['bp'].plot.box(figsize=(16, 5))
plt.show()推论:在上图中,可以看到血压的分布,并且在子图中可以看到 bp 列中有一些异常值。
将分类值(对象)转换为分类值(int)
data['classification'] = data['classification'].map({'ckd':1,'notckd':0})
data['htn'] = data['htn'].map({'yes':1,'no':0})
data['dm'] = data['dm'].map({'yes':1,'no':0})
data['cad'] = data['cad'].map({'yes':1,'no':0})
data['appet'] = data['appet'].map({'good':1,'poor':0})
data['ane'] = data['ane'].map({'yes':1,'no':0})
data['pe'] = data['pe'].map({'yes':1,'no':0})
data['ba'] = data['ba'].map({'present':1,'notpresent':0})
data['pcc'] = data['pcc'].map({'present':1,'notpresent':0})
data['pc'] = data['pc'].map({'abnormal':1,'normal':0})
data['rbc'] = data['rbc'].map({'abnormal':1,'normal':0})
data['classification'].value_counts()分析相关性
plt.figure(figsize = (19,19))
sns.heatmap(df.corr(), annot = True, cmap = 'coolwarm') # looking for strong correlations with "class" row探索性数据分析 (EDA)
data.shapedata.columns删除空值
(data.shape[0],data.dropna().shape[0])分析: 数据集中有 158 个空值。
有两种处理方法:
(1)删除所有空值;(2)删除 NA 值时保留它们
因为数据集不是那么大,如果删除空值,则向机器学习模型提供的数据非常少,那么性能会非常低,并且所删除的数据可能会影响其他特征。
-->保留数据
data.dropna(inplace=True)
data.shape构建模型
逻辑回归
from sklearn.linear_model import LogisticRegressionlogreg = LogisticRegression()
X = data.iloc[:,:-1]
y = data['classification']
X_train, X_test, y_train, y_test = train_test_split(X,y, stratify = y, shuffle = True)
logreg.fit(X_train,y_train)训练精确度
logreg.score(X_train,y_train)测试精确度
logreg.score(X_test,y_test)变量系数
从逻辑回归中读取系数的示例:年龄增加一单位使个体患 CKD 的可能性约为 e^0.14 倍,而血压增加一单位使个体约 e^-0.07 倍可能患有 CKD。
pd.DataFrame(logreg.coef_, columns=X.columns)Confusion Matrix
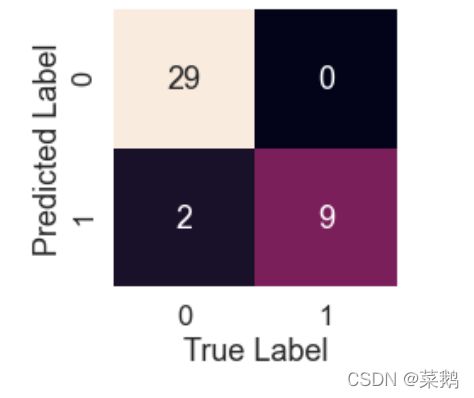
sns.set(font_scale=1.5)def plot_conf_mat(y_test,y_preds):
"""
This function will be heloing in plotting the confusion matrix by using seaborn
"""
fig,ax=plt.subplots(figsize=(3,3))
ax=sns.heatmap(confusion_matrix(y_test,y_preds),annot=True,cbar=False)
plt.xlabel("True Label")
plt.ylabel("Predicted Label")log_pred = logreg.predict(X_test)
plot_conf_mat(y_test, log_pred)K-最近邻分类器
df["classification"].value_counts()balance_df = pd.concat([df[df["classification"] == 0], df[df["classification"] == 1].sample(n = 115, replace = True)], axis = 0)
balance_df.reset_index(drop=True, inplace=True)
balance_df["classification"].value_counts()缩放数据
ss = StandardScaler()
ss.fit(X_train)
X_train = ss.transform(X_train)
X_test = ss.transform(X_test)调整 KNN 模型以获得更好的准确性
from sklearn.neighbors import KNeighborsClassifierknn = KNeighborsClassifier()
params = {
"n_neighbors":[3,5,7,9],
"weights":["uniform","distance"],
"algorithm":["ball_tree","kd_tree","brute"],
"leaf_size":[25,30,35],
"p":[1,2]
}
gs = GridSearchCV(knn, param_grid=params)
model = gs.fit(X_train,y_train)
preds = model.predict(X_test)
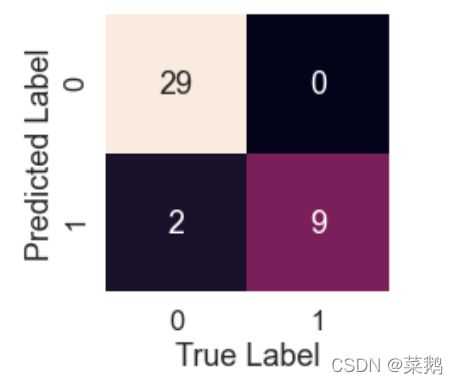
accuracy_score(y_test, preds)KNN模型的混淆矩阵
knn_pred = model.predict(X_test)
plot_conf_mat(y_test, knn_pred)tn, fp, fn, tp = confusion_matrix(y_test, preds).ravel()
print(f'True Neg: {tn}')
print(f'False Pos: {fp}')
print(f'False Neg: {fn}')
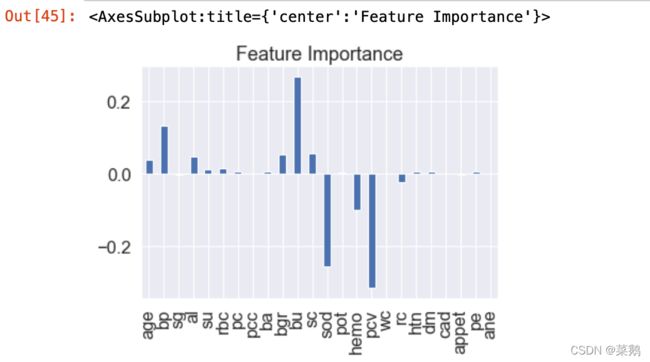
print(f'True Pos: {tp}')特征重要性
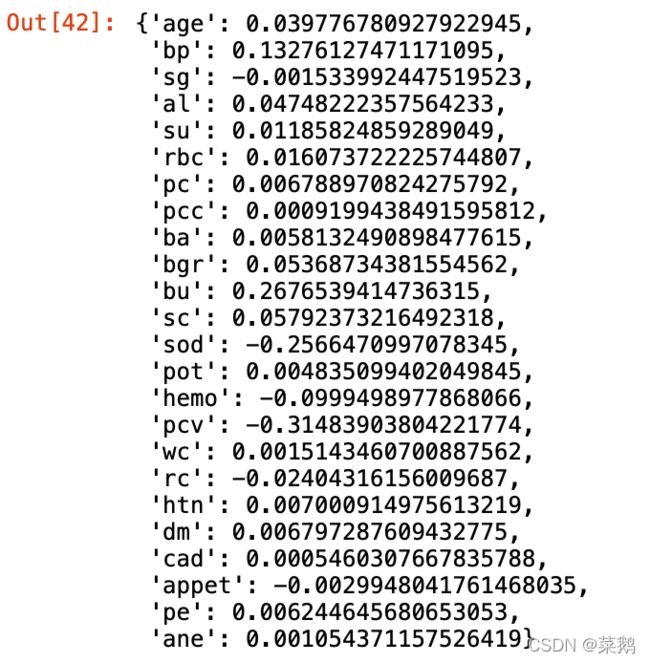
feature_dict=dict(zip(df.columns,list(logreg.coef_[0])))
feature_dict可视化特征重要性
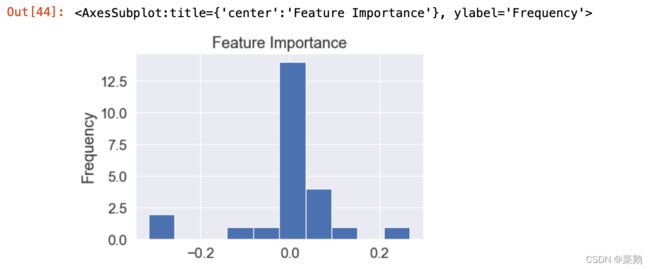
feature_df=pd.DataFrame(feature_dict,index=[0])
feature_df.T.plot(kind="hist",legend=False,title="Feature Importance")可视化特征重要性——转置
feature_df=pd.DataFrame(feature_dict,index=[0])
feature_df.T.plot(kind="bar",legend=False,title="Feature Importance")保存模型
import pickle
# Now with the help of pickle model we will be saving the trained model
saved_model = pickle.dumps(logreg)
# Load the pickled model
logreg_from_pickle = pickle.loads(saved_model)
# Now here we will load the model
logreg_from_pickle.predict(X_test)原文链接:Predicting Chronic Kidney Disease using Machine Learning