基于R语言利用CIBERSORT分析免疫浸润(一)
第一步:认识CIBERSOR
首先我们认识到了单细胞当中每个细胞的特征表达不同。因此对于Bulk数据而言,多个细胞的表达,应该就是这些单个细胞表达的一种加合,我们就可以利用单个细胞的特征使用线性去卷积方法去计算Bulk数据中单细组分含量。
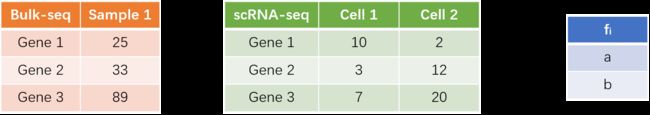
如图,我们可以认为在Sample 1样品中,Cell 1组分含量为a,Cell 2的组分含量为 b。
第二步:CIBERSORT算法
整个算法代码如下,除了此处复制粘贴外,也可以通过网站:https://rdrr.io/github/singha53/amritr/src/R/supportFunc_cibersort.R获取该代码。
补充:也可以使用CIBERSORT的在线工具。
#' CIBERSORT R script v1.03 (last updated 07-10-2015)
#' Note: Signature matrix construction is not currently available; use java version for full functionality.
#' Author: Aaron M. Newman, Stanford University ([email protected])
#' Requirements:
#' R v3.0 or later. (dependencies below might not work properly with earlier versions)
#' install.packages('e1071')
#' install.pacakges('parallel')
#' install.packages('preprocessCore')
#' if preprocessCore is not available in the repositories you have selected, run the following:
#' source("http://bioconductor.org/biocLite.R")
#' biocLite("preprocessCore")
#' Windows users using the R GUI may need to Run as Administrator to install or update packages.
#' This script uses 3 parallel processes. Since Windows does not support forking, this script will run
#' single-threaded in Windows.
#'
#' Usage:
#' Navigate to directory containing R script
#'
#' In R:
#' source('CIBERSORT.R')
#' results <- CIBERSORT('sig_matrix_file.txt','mixture_file.txt', perm, QN)
#'
#' Options:
#' i) perm = No. permutations; set to >=100 to calculate p-values (default = 0)
#' ii) QN = Quantile normalization of input mixture (default = TRUE)
#'
#' Input: signature matrix and mixture file, formatted as specified at http://cibersort.stanford.edu/tutorial.php
#' Output: matrix object containing all results and tabular data written to disk 'CIBERSORT-Results.txt'
#' License: http://cibersort.stanford.edu/CIBERSORT_License.txt
#' Core algorithm
#' @param X cell-specific gene expression
#' @param y mixed expression per sample
#' @export
CoreAlg <- function(X, y){
#try different values of nu
svn_itor <- 3
res <- function(i){
if(i==1){nus <- 0.25}
if(i==2){nus <- 0.5}
if(i==3){nus <- 0.75}
model<-e1071::svm(X,y,type="nu-regression",kernel="linear",nu=nus,scale=F)
model
}
if(Sys.info()['sysname'] == 'Windows') out <- parallel::mclapply(1:svn_itor, res, mc.cores=1) else
out <- parallel::mclapply(1:svn_itor, res, mc.cores=svn_itor)
nusvm <- rep(0,svn_itor)
corrv <- rep(0,svn_itor)
#do cibersort
t <- 1
while(t <= svn_itor) {
weights = t(out[[t]]$coefs) %*% out[[t]]$SV
weights[which(weights<0)]<-0
w<-weights/sum(weights)
u <- sweep(X,MARGIN=2,w,'*')
k <- apply(u, 1, sum)
nusvm[t] <- sqrt((mean((k - y)^2)))
corrv[t] <- cor(k, y)
t <- t + 1
}
#pick best model
rmses <- nusvm
mn <- which.min(rmses)
model <- out[[mn]]
#get and normalize coefficients
q <- t(model$coefs) %*% model$SV
q[which(q<0)]<-0
w <- (q/sum(q))
mix_rmse <- rmses[mn]
mix_r <- corrv[mn]
newList <- list("w" = w, "mix_rmse" = mix_rmse, "mix_r" = mix_r)
}
#' do permutations
#' @param perm Number of permutations
#' @param X cell-specific gene expression
#' @param y mixed expression per sample
#' @export
doPerm <- function(perm, X, Y){
itor <- 1
Ylist <- as.list(data.matrix(Y))
dist <- matrix()
while(itor <= perm){
#print(itor)
#random mixture
yr <- as.numeric(Ylist[sample(length(Ylist),dim(X)[1])])
#standardize mixture
yr <- (yr - mean(yr)) / sd(yr)
#run CIBERSORT core algorithm
result <- CoreAlg(X, yr)
mix_r <- result$mix_r
#store correlation
if(itor == 1) {dist <- mix_r}
else {dist <- rbind(dist, mix_r)}
itor <- itor + 1
}
newList <- list("dist" = dist)
}
#' Main functions
#' @param sig_matrix file path to gene expression from isolated cells
#' @param mixture_file heterogenous mixed expression
#' @param perm Number of permutations
#' @param QN Perform quantile normalization or not (TRUE/FALSE)
#' @export
CIBERSORT <- function(sig_matrix, mixture_file, perm=0, QN=TRUE){
#read in data
X <- read.table(sig_matrix,header=T,sep="\t",row.names=1,check.names=F)
Y <- read.table(mixture_file, header=T, sep="\t", row.names=1,check.names=F)
X <- data.matrix(X)
Y <- data.matrix(Y)
#order
X <- X[order(rownames(X)),]
Y <- Y[order(rownames(Y)),]
P <- perm #number of permutations
#anti-log if max < 50 in mixture file
if(max(Y) < 50) {Y <- 2^Y}
#quantile normalization of mixture file
if(QN == TRUE){
tmpc <- colnames(Y)
tmpr <- rownames(Y)
Y <- preprocessCore::normalize.quantiles(Y)
colnames(Y) <- tmpc
rownames(Y) <- tmpr
}
#intersect genes
Xgns <- row.names(X)
Ygns <- row.names(Y)
YintX <- Ygns %in% Xgns
Y <- Y[YintX,]
XintY <- Xgns %in% row.names(Y)
X <- X[XintY,]
#standardize sig matrix
X <- (X - mean(X)) / sd(as.vector(X))
#empirical null distribution of correlation coefficients
if(P > 0) {nulldist <- sort(doPerm(P, X, Y)$dist)}
#print(nulldist)
header <- c('Mixture',colnames(X),"P-value","Correlation","RMSE")
#print(header)
output <- matrix()
itor <- 1
mixtures <- dim(Y)[2]
pval <- 9999
#iterate through mixtures
while(itor <= mixtures){
y <- Y[,itor]
#standardize mixture
y <- (y - mean(y)) / sd(y)
#run SVR core algorithm
result <- CoreAlg(X, y)
#get results
w <- result$w
mix_r <- result$mix_r
mix_rmse <- result$mix_rmse
#calculate p-value
if(P > 0) {pval <- 1 - (which.min(abs(nulldist - mix_r)) / length(nulldist))}
#print output
out <- c(colnames(Y)[itor],w,pval,mix_r,mix_rmse)
if(itor == 1) {output <- out}
else {output <- rbind(output, out)}
itor <- itor + 1
}
#save results
write.table(rbind(header,output), file="CIBERSORT-Results.txt", sep="\t", row.names=F, col.names=F, quote=F)
#return matrix object containing all results
obj <- rbind(header,output)
obj <- obj[,-1]
obj <- obj[-1,]
obj <- matrix(as.numeric(unlist(obj)),nrow=nrow(obj))
rownames(obj) <- colnames(Y)
colnames(obj) <- c(colnames(X),"P-value","Correlation","RMSE")
obj
}关于算法,我们可以从这些代码中看出CoreAlg,doPerm和CIBERSORT为核心的自定义函数,整个算法是基于线性支持向量回归(SVR),即进行SVM反卷积推算。
注:SVM和SVR虽然原理类似,但SVM是用作分类,SVR才是用于回归。
理解了整个代码后,我们将其保存为R脚本文件,命名为“Cibersort.R”。方便后续使用。
第三步:准备数据集(LM22.txt和表达谱数据)
LM22.txt数据集是基准数据库文件,表示22种免疫细胞的marker基因,可以从网站:Robust enumeration of cell subsets from tissue expression profiles | Nature Methods。其中Supplementary information有Supplementary Table 1链接(Leukocyte signature matrix (LM22). Details of LM22, including gene expression matrix and source data. (XLS 424 kb)),成功下载后是.csv文件。我们只需要其中免疫细胞marker基因表达数据,将其提取出来保存为“LM22.txt”文件即可。
至于表达谱数据,进行前期数据清洗,如去除表达量低的基因等,即可用于分析。把处理后的表达谱数据导出为.txt文件,命名为“Data.txt”。
注:CIBERSORT种自带log转换功能,当然也可以对自己的数据集转换。
第四步(最后一步):CIBERSORT分析
首先加载R脚本文件,然后使用CIBERSORT函数。具体代码如下:
source("Cibersort.R")
result <- CIBERSORT("LM22.txt", "Data.txt", perm = 1000, QN = TRUE)
#perm为置换次数。用于估算单个样本免疫浸润的p值。
#QN为分位数归一化。一般芯片数据设置为T,测序数据设置为F。
注:CIBERSORT算法中自带输出结果代码,默认保存结果文件名为“CIBERSORT-Results.txt”。
到此,关于免疫浸润分析就结束了,关于分析结果的可视化后续进行!