基于R语言利用CIBERSORT分析免疫浸润(二)
引言:本节重点
(1)在我写(一)的时候,很多小伙伴相信直接使用遇到了很多问题,本节就主要遇到问题进行回答:
①R包版本问题
②Data数据格式问题
③CIBERSORT官方代码问题
并将附上全部代码
(2)此外,本节将介绍分析后的数据常见的可视化方式
一、问题解决
1、R包版本问题
相信很多人都会遇到这个问题。如“这个R包版本不适用于该版本R”、“连接错误”、“依赖包安装无效或版本不足”等等。这时候通常解决办法两种:
(1)重装R和Studio,但这样就显得过于麻烦了。
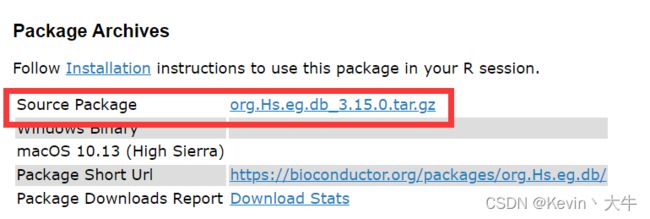
(2)相信第二种办法很多人就愿意去尝试了。一个是利用R官网提供的R包下载路径,通过自己下载tar.zip文件导入R中;二是在BiocConductor官网下载R包的tar.zip文件,同样导入R中,我通常采用的该方法。(该方法将在第二个问题进行一个图示讲解)
2、Data数据格式问题
很多人使用CIBERSORT方法的时候,是没有注意Data和LM22两套数据的,因此极其容易发生错误,最需要注意的地方之一就是LM22的行名是ENTREZ ID,如果Data的行名是GENE SYMBOL是不是就无法进行计算了呢?答案是肯定的,所以我们就需要将Data的行名改为ENTREZ ID。具体操作如下
①下载并导入安装包(因为R无法使用install.packages和BiocManager::install下载)
https://www.bioconductor.org/packages/release/data/annotation/html/org.Hs.eg.db.html
当然,这一个是不够的,你需要下载这么多
②使用以下代码替换GENE SYMBOL
library(org.Hs.eg.db) library(clusterProfiler) gene.df <- bitr(rownames(data), fromType = "SYMBOL", toType = c("ENTREZID"), OrgDb = org.Hs.eg.db) data <- data[which(rownames(data) %in% gene.df[,1]),] rownames(data) <- gene.df[,2]3、CIBERSORT官方代码问题
第一个问题是CIBERSORT没有考虑到LM22数据中有NA的问题,因此会导致运行的时候R直接挂掉。所以我们需要在官方代码的主函数CIBERSORT()中添加这句话。
第二个问题就很奇怪了,如果调用CIBERSORT()这个函数总是会报Model is empty!但是在我一句话一句话检查的时候就没有,因此我直接把CIBERSORT()主函数去掉,咱们直接一句一句运行即可。最终所有代码如下
library(dplyr) library(limma) setwd("D:\\工作文件\\CSDN\\CINBERSORT(x)\\示例数据") rm(list = ls()) ####读入数据#### #表达谱数据 data <- read.table(".\\GSE数据\\GSE159661_series_matrix.txt", sep = "\t", comment.char = "!", fill = T) colnames(data) <- data[1,] data <- data[-1,] #注释数据 meta <- read.table(".\\GPL数据\\GPL21185-21174.txt", sep = "\t", comment.char = "#", fill = T) colnames(meta) <- meta[1,] meta <- meta[-1,] ####注释数据#### #合并信息 colnames(data)[1] <- "ID" meta <- meta[,c(1,6)] data <- merge(data, meta, by = "ID") data <- data[!is.na(data$GENE_SYMBOL),] data <- data[data$GENE_SYMBOL != "",] #相同基因取均值 GENE_SYMBOL <- data$GENE_SYMBOL data <- data[,-c(1,14)] data <- lapply(data, as.numeric) %>% as.data.frame(.) data <- aggregate(.~GENE_SYMBOL,data = data, mean) rownames(data) <- data[,1] data <- data[,-1] #对照信息 group <- c("sensitive","resistant","resistant", "resistant","sensitive","resistant", "sensitive","resistant","resistant", "resistant","sensitive","resistant") %>% factor(, levels = c("resistant","sensitive"), ordered = F) #Entrez id信息 library(org.Hs.eg.db) library(clusterProfiler) gene.df <- bitr(rownames(data), fromType = "SYMBOL", toType = c("ENTREZID"), OrgDb = org.Hs.eg.db) data <- data[which(rownames(data) %in% gene.df[,1]),] rownames(data) <- gene.df[,2] ####数据预处理#### #log2转换 # data <- log2(data) ####差异分析#### group <- model.matrix(~factor(group)+0) colnames(group) <- c("resistant","sensitive") df.fit <- lmFit(data, group) df.matrix <- makeContrasts(resistant - sensitive, levels = group) fit <- contrasts.fit(df.fit, df.matrix) fit <- eBayes(fit) tempOutput <- topTable(fit, n = Inf, adjust = "fdr") diffGene <- rownames(tempOutput)[which(abs(tempOutput$logFC) > 1 & tempOutput$adj.P.Val < 0.05)] # data <- data[(rownames(data) %in% diffGene),] write.table(data, "Data.txt", sep = "\t", row.names = T, col.names = T) ####Cibersort分析#### rm(list = ls()) source("D:\\工作文件\\CSDN\\CINBERSORT(x)\\CIBERSORT.R") result <- CIBERSORT() rm(list = ls()) ####Cibersort分析#### #' @param X cell-specific gene expression #' @param y mixed expression per sample #' @export CoreAlg <- function(X, y){ #try different values of nu svn_itor <- 3 res <- function(i){ if(i==1){nus <- 0.25} if(i==2){nus <- 0.5} if(i==3){nus <- 0.75} model<-e1071::svm(X,y,type="nu-regression",kernel="linear",nu=nus,scale=F) model } if(Sys.info()['sysname'] == 'Windows') out <- parallel::mclapply(1:svn_itor, res, mc.cores=1) else out <- parallel::mclapply(1:svn_itor, res, mc.cores=svn_itor) nusvm <- rep(0,svn_itor) corrv <- rep(0,svn_itor) #do cibersort t <- 1 while(t <= svn_itor) { weights = t(out[[t]]$coefs) %*% out[[t]]$SV weights[which(weights<0)]<-0 w<-weights/sum(weights) u <- sweep(X,MARGIN=2,w,'*') k <- apply(u, 1, sum) nusvm[t] <- sqrt((mean((k - y)^2))) corrv[t] <- cor(k, y) t <- t + 1 } #pick best model rmses <- nusvm mn <- which.min(rmses) model <- out[[mn]] #get and normalize coefficients q <- t(model$coefs) %*% model$SV q[which(q<0)]<-0 w <- (q/sum(q)) mix_rmse <- rmses[mn] mix_r <- corrv[mn] newList <- list("w" = w, "mix_rmse" = mix_rmse, "mix_r" = mix_r) } #' do permutations #' @param perm Number of permutations #' @param X cell-specific gene expression #' @param y mixed expression per sample #' @export doPerm <- function(perm, X, Y){ itor <- 1 Ylist <- as.list(data.matrix(Y)) dist <- matrix() while(itor <= perm){ #print(itor) #random mixture yr <- as.numeric(Ylist[sample(length(Ylist),dim(X)[1])]) #standardize mixture yr <- (yr - mean(yr)) / sd(yr) #run CIBERSORT core algorithm result <- CoreAlg(X, yr) mix_r <- result$mix_r #store correlation if(itor == 1) {dist <- mix_r} else {dist <- rbind(dist, mix_r)} itor <- itor + 1 } newList <- list("dist" = dist) } #' Main functions #' @param sig_matrix file path to gene expression from isolated cells #' @param mixture_file heterogenous mixed expression #' @param perm Number of permutations #' @param QN Perform quantile normalization or not (TRUE/FALSE) #' @export perm = 999 QN = T #read in data X <- read.table("D:\\工作文件\\CSDN\\CINBERSORT(x)\\LM22\\LM22.txt",header=T,sep="\t",row.names=1,check.names=F) Y <- read.table("D:\\工作文件\\CSDN\\CINBERSORT(x)\\示例数据\\Data.txt", header=T, sep="\t", row.names=1,check.names=F) #去除NA X <- na.omit(X) X <- data.matrix(X) Y <- data.matrix(Y) #order X <- X[order(rownames(X)),] Y <- Y[order(rownames(Y)),] P <- perm #number of permutations #anti-log if max < 50 in mixture file if(max(Y) < 50) {Y <- 2^Y} #quantile normalization of mixture file if(QN == TRUE){ tmpc <- colnames(Y) tmpr <- rownames(Y) Y <- preprocessCore::normalize.quantiles(Y) colnames(Y) <- tmpc rownames(Y) <- tmpr } #intersect genes Xgns <- row.names(X) Ygns <- row.names(Y) YintX <- Ygns %in% Xgns Y <- Y[YintX,] XintY <- Xgns %in% row.names(Y) X <- X[XintY,] #standardize sig matrix X <- (X - mean(X)) / sd(as.vector(X)) #empirical null distribution of correlation coefficients if(P > 0) {nulldist <- sort(doPerm(P, X, Y)$dist)} #print(nulldist) header <- c('Mixture',colnames(X),"P-value","Correlation","RMSE") #print(header) output <- matrix() itor <- 1 mixtures <- dim(Y)[2] pval <- 9999 #iterate through mixtures while(itor <= mixtures){ y <- Y[,itor] #standardize mixture y <- (y - mean(y)) / sd(y) #run SVR core algorithm result <- CoreAlg(X, y) #get results w <- result$w mix_r <- result$mix_r mix_rmse <- result$mix_rmse #calculate p-value if(P > 0) {pval <- 1 - (which.min(abs(nulldist - mix_r)) / length(nulldist))} #print output out <- c(colnames(Y)[itor],w,pval,mix_r,mix_rmse) if(itor == 1) {output <- out} else {output <- rbind(output, out)} itor <- itor + 1 } #save results write.table(rbind(header,output), file="CIBERSORT-Results.txt", sep="\t", row.names=F, col.names=F, quote=F) #return matrix object containing all results obj <- rbind(header,output) obj <- obj[,-1] obj <- obj[-1,] obj <- matrix(as.numeric(unlist(obj)),nrow=nrow(obj)) rownames(obj) <- colnames(Y) colnames(obj) <- c(colnames(X),"P-value","Correlation","RMSE") obj
二、数据可视化
这里主要讲解两个比较常用的可视化呈现方式,第一种是基于对照的箱线图(ggplot2包),第二种是基于组成的直方图(ggpubr包)。这里采用最粗暴的讲解方式:代码+图示结果!
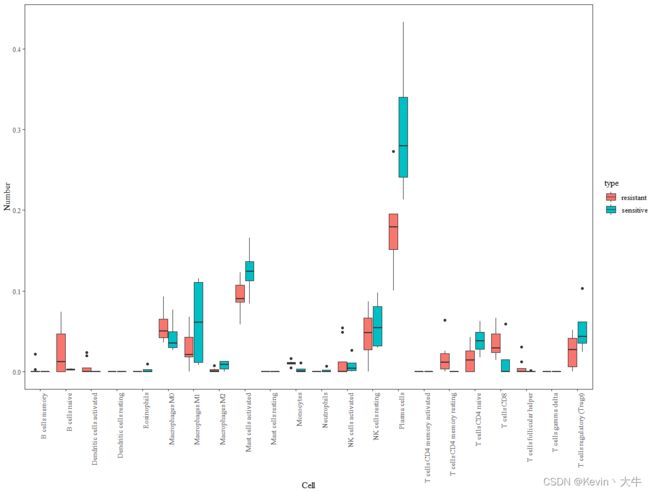
1、箱线图:主要用到的函数是ggplot,废话不多说,代码和结果如下
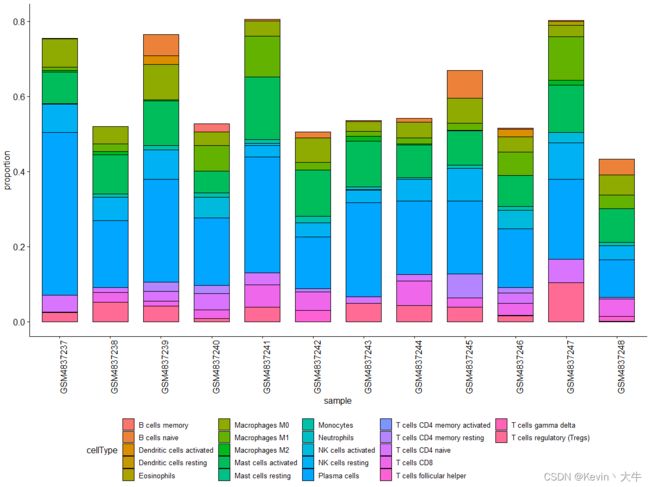
library(ggplot2) library(ggpubr) ####数据可视化#### group <- c("sensitive","resistant","resistant", "resistant","sensitive","resistant", "sensitive","resistant","resistant", "resistant","sensitive","resistant") %>% factor(, levels = c("resistant","sensitive"), ordered = F) obj <- as.data.frame(obj) obj <- cbind(obj, group) obj$Sample <- rownames(obj) cell <- data.frame(type = "", cellType = "", proportion = 0) for(i in 3:24){ part <- aggregate(obj[,i], list(obj$group), function(x){x})[,2] for(m in part[[1]]){ cell <- rbind(cell, data.frame(type = "resistant", cellType = colnames(obj)[i], proportion = m)) } for(n in part[[2]]){ cell <- rbind(cell, data.frame(type = "sensitive", cellType = colnames(obj)[i], proportion = n)) } } cell <- cell[-1,] ggplot(cell, aes(x = cellType, y = proportion, fill = type))+ geom_boxplot()+ ggtitle(NULL)+ labs(x = "Cell", y = "Number")+ theme_set(theme_bw())+ theme(panel.grid.major = element_line(colour = NA), panel.grid.minor = element_blank(), text=element_text(size = 12, family = "serif"), axis.text.x = element_text(angle = 90, hjust = 1))2、直方图:主要函数是ggbarplot。
sample <- data.frame(sample = "", proportion = 0, cellType = "") for(i in 3:24){ part <- aggregate(obj[,i], list(obj$Sample), function(x){x}) part$cellType <- colnames(obj)[i] colnames(part) <- c("sample", "proportion", "cellType") sample <- rbind(sample, part) } sample <- sample[-1,] ggbarplot(sample, x = "sample", y= "proportion", fill = "cellType")+ theme(axis.text.x = element_text(angle = 90,hjust = 1,vjust = 1,size = 12), legend.position = "bottom")