用于深度学习的医学图像预处理
用于深度学习的医学图像数据,往往非常庞大,如果从网上下载公开数据集数据,往往有几十GB的图象数据,我们需要先进行预处理,将其转换成适合深度学习网络训练的形式:
或者我们还需要:Dicom变为nii,变为JPG;去除扫描床影响;为每个图象重命名等等。
一、文件批量重命名
1、文件夹重命名:
import os,sys
def File_Rename(path):
filelist = os.listdir(path)
total_num = len(filelist)
i = 0
for file_name in filelist:
os.rename((path + file_name), (path + str(i).zfill(2))) # 子文件夹重命名
print(file_name, "has been renamed successfully! New name is: ",str(i).zfill(2))
i = i + 1
if __name__ == '__main__':
path = r'E:/DeepLearningWorkSpace/PyCharmWorkSpace/Test/Bo_Test/thing/'
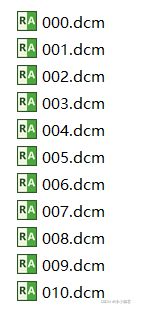
File_Rename(path) #调用定义的函数2、文件重命名:dicom文件为例
import os
def rename(path):
filelist = os.listdir(path)
total_num = len(filelist)
i = 0
for item in filelist:
if item.endswith('.dcm'):
src = os.path.join(os.path.abspath(path), item)
dst = os.path.join(os.path.abspath(path), str(i).zfill(3) + '.dcm')
os.rename(src, dst)
print(item, "has been renamed successfully! New name is: ", str(i).zfill(3) + '.dcm')
i = i + 1
print('total %d to rename & converted %d dcms' % (total_num, i))
if __name__ == '__main__':
path = r'E:/DeepLearningWorkSpace/PyCharmWorkSpace/Test/Bo_Test/thing/00/'
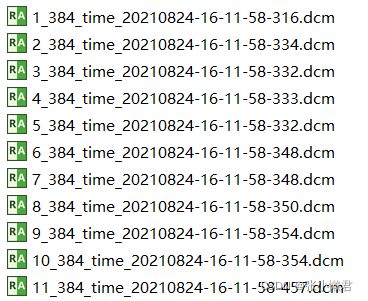
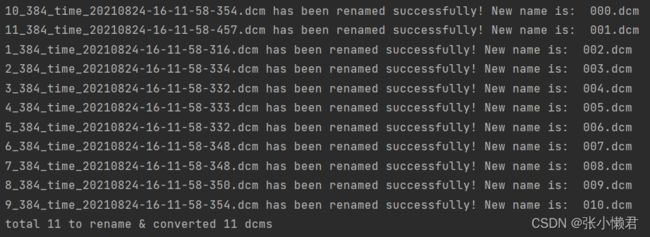
rename(path)但是这里,出现了一个问题:没有按照顺序命名,它将倒数第二各10_384_time.....命名成000,但我想将1_384_time....命名为000,然后依次往下。
(这里明白了,计算机是按照第一位,第二位,这样一位一位识别的,10_,11_,1_,先识别10_,然后是11_,和1_。所以以后对文件命名时,尽量写成001,002.....010。而且再重命名前做好备份,以防万一。)
现在遇到上面这种情况,可以:
获取文件名 中:_384前面的内容,再命名。
file_name = '10_384_time_20210824-16-11-58-316.dcm'
print(file_name[0:file_name.rfind('_384')])
>>103、 批量文件夹内文件重命名:
import os
def rename(path):
filelist = os.listdir(path)
total_num = len(filelist)
i = 0
for item in filelist:
if item.endswith('.dcm'):
src = os.path.join(os.path.abspath(path), item)
dst = os.path.join(os.path.abspath(path), str(i)zfill(3) + '.dcm')
os.rename(src, dst)
print('converting %s to %s ...' % (src, dst))
i = i + 1
print('total %d to rename & converted %d dcms' % (total_num, i))
Dir_pathes = 'E:/DeepLearningWorkSpace/PyCharmWorkSpace/date/NBIA_lung/CBCT'
Dir_list = os.listdir(Dir_pathes)
Dir_num = len(Dir_list)
for k in Dir_list:
path = os.path.join(Dir_pathes,k)
rename(path)然后对文件重命名结束之后,可以对dicom文件进行处理。
二、移除扫描床处理
Fuction_quchuang.py
import matplotlib.pyplot as plt
import numpy as np
import pydicom
import cv2
import SimpleITK as sitk
from PIL import Image
from skimage.morphology import disk, rectangle, binary_dilation, binary_erosion, binary_closing, binary_opening
def read_dicom_data(file_name):
file = sitk.ReadImage(file_name)
data = sitk.GetArrayFromImage(file)
print(data.shape)
data = np.squeeze(data, axis=0)
print(data.shape)
data = np.int32(data)
dicom_dataset = pydicom.dcmread(file_name)
slice_location = dicom_dataset.SliceLocation #获取层间距
return data, data.shape[0], data.shape[1], slice_location
def window(window, img_data):
if window == 'Lung':
img_data[img_data < -1150] = -1150
img_data[img_data > 350] = 350
elif window == 'Pelvic':
img_data[img_data < -138] = -138
img_data[img_data > 238] = 238
elif window == 'Chest':
img_data[img_data < -160] = -160
img_data[img_data > 240] = 240
elif window == 'Chest_scatter':
img_data[img_data < -752] = -752
img_data[img_data > 838] = 838
elif window == 'Pelvic_scatter':
img_data[img_data < -300] = -300
img_data[img_data > 240] = 240
else:
img_data[img_data < 0] = 0
img_data[img_data > 80] = 80
return img_data
def find_max_region(mask_sel):
contours, hierarchy = cv2.findContours(mask_sel, cv2.RETR_TREE, cv2.CHAIN_APPROX_NONE)
# 找到最大区域并填充
area = []
for j in range(len(contours)):
area.append(cv2.contourArea(contours[j]))
max_idx = np.argmax(area)
max_area = cv2.contourArea(contours[max_idx])
for k in range(len(contours)):
if k != max_idx:
cv2.fillPoly(mask_sel, [contours[k]], 0)
return mask_sel
def quchuang(dcm_path,window1,window2):
# 读入dicom图像
dcm_path = dcm_path
pixel_array, rows, columns, slice_location = read_dicom_data(dcm_path)
pixel_array1 = pixel_array.copy()
pixel_array2 = pixel_array.copy()
pixel_array1 = window(window1, pixel_array1)
imageData1 = (pixel_array1 - pixel_array1.min()) * 255.0 / (pixel_array1.max() - pixel_array1.min())
imageData1 = np.uint8(imageData1)
pixel_array2 = window(window2, pixel_array2)
imageData2 = (pixel_array2 - pixel_array2.min()) * 255.0 / (pixel_array2.max() - pixel_array2.min())
imageData2 = np.uint8(imageData2)
# 二值化
ret, binary = cv2.threshold(imageData2, 3, 255, cv2.THRESH_BINARY)
# 腐蚀
selem = disk(3)
fushi = binary_erosion(binary, selem)
# 找最大连通区域
binary = np.uint8(fushi)
max_region = find_max_region(binary)
# 膨胀
selem = disk(3)
pengzhang = binary_dilation(max_region, selem)
#最左最右边两列像素值为0
pengzhang[:, 0] = 0
pengzhang[:, 511] = 0
#填充
A = np.uint8(pengzhang)
h, w = A.shape[:2]
#print(pengzhang.shape[:2])
mask_tp = np.zeros((h + 2, w + 2), np.uint8)
temp = A.copy()
temp2 = A.copy()
cv2.floodFill(temp, mask_tp, (1, 1), 255)
cv2.floodFill(temp2, mask_tp, (w - 2, h - 2), 255)
rt = cv2.bitwise_not(temp)
Tianchong = rt
Tianchong[rt > 0] = 255
image_process = np.uint8((Tianchong / 255) * imageData1)
return image_process
if __name__ == '__main__':
dcm_path = "E:/DeepLearningWorkSpace/PyCharmWorkSpace/date/CBCT_Pancreatic/22/CT/22/24.dcm"
window1 = 'Pelvic'
window2 = 'Pelvic_scatter'
image_array = quchuang(dcm_path, window1, window2)
#image = Image.fromarray(image_array)
save_path = 'E:/DeepLearningWorkSpace/PyCharmWorkSpace/date/CBCT_Pancreatic/JPG/buchong'
cv2.imwrite(save_path + '/22_24.jpg', image_array)以上是处理一幅dicom并将其保存为JPG格式的代码。
from Function_quchuang import read_dicom_data,window,find_max_region,quchuang #调用上一个代码函数
import os
import cv2
import re
Dir_pathes = 'E:/DeepLearningWorkSpace/PyCharmWorkSpace/date/Pancreatic/22/CT/'
Dir_list = os.listdir(Dir_pathes)
Dir_num = len(Dir_list)
for k in Dir_list:
path = os.path.join(Dir_pathes, k)
dir_list = os.listdir(path)
dir_num = len(dir_list)
for j in dir_list:
window1 = 'Pelvic'
window2 = 'Pelvic_scatter'
image_array = quchuang(path + '/' + j, window1, window2)
save_path = 'E:/DeepLearningWorkSpace/PyCharmWorkSpace/date/Pancreatic/JPG/train_B'
cv2.imwrite(save_path + '/' + k + '_'+ re.sub("\D", "", j) +'.jpg', image_array)
print('%s libingren, di %s fu image' % (k, j))以上是批量处理移除扫描床的代码.
一些去床的具体内容在: DICOM的理解与学习2_张小懒君的博客-CSDN博客