limma | 分层样本的差异分析这样搞!(二)

1写在前面
上期介绍了用limma包做配对样本的差异分析。
本期介绍一下Multi-level如何处理吧。
应用场景:
Control和Diseased的T细胞和B细胞分层对比。
2用到的包
rm(list = ls())
library(tidyverse)
library(limma)
library(GEOquery)
3示例数据
这里我们还是利用上期介绍的GEO数据库上的dataset。
在3个样本中对T细胞和B细胞分别进行了转录组分析。
每个样本的细胞都分为Control或anti-BTLA组。
GSE194314 <- getGEO('GSE194314', destdir=".",getGPL = F)
exprSet <- exprs(GSE194314[[1]])
exprSet <- normalizeBetweenArrays(exprSet) %>%
log2(.)

4获取分组数据
pdata <- pData(GSE194314[[1]])

5整理分组数据
这里我们提取出分组数据后转为factor。
individuals <- factor(unlist(lapply(pdata$characteristics_ch1.1,function(x) strsplit(as.character(x),": ")[[1]][2])))
cell_type <- factor(unlist(lapply(pdata$characteristics_ch1,function(x) strsplit(as.character(x),": ")[[1]][2])))
cell_type <- gsub("[[:punct:]]","", cell_type)
cell_type <- gsub("\\s","_", cell_type)
treatment <- unlist(lapply(pdata$characteristics_ch1.2,function(x) strsplit(as.character(x),": ")[[1]][2]))
treatment <- factor(treatment,levels = unique(treatment))
treatment<- gsub("-", "_", treatment)

6Multi-level的处理与理解
6.1 整理分组矩阵
如果我们的目的是比较两种细胞类型间的差异,可以在样本内部进行,因为每个样本都会产生两种细胞类型的值。
如果我们想将Control与anti-BTLA进行比较,这种比较就是在不同样本之间进行了。
这里大家可以理解为,需要进行组内和组间比较,处理样本时需要用到random effect,在limma包中需要调用duplicateCorrelation函数进行处理。
measures <- factor(paste(treatment, cell_type,sep="."))
design <- model.matrix(~ 0 + measures)
colnames(design) <- levels(measures)

6.2 相关性计算
然后,我们在同一样本上计算不同组学间的相关性。
corfit <- duplicateCorrelation(exprSet, design, block = individuals)
corfit$consensus
6.3 模型拟合
fit <- lmFit(exprSet, design, block = individuals, correlation = corfit$consensus)
6.4 设置比较组
这里我们设置一下比较组,用-分隔。
cm <- makeContrasts(antiBLTA_vs_Control_For_Bcell =
anti_BTLA.Purified_B_cell-Control.Purified_B_cell,
antiBLTA_vs_Control_For_Tcell =
anti_BTLA.Purified_CD4_T_cell-Control.Purified_CD4_T_cell,
Bcell_vs_Tcell_For_Control =
Control.Purified_B_cell-Control.Purified_CD4_T_cell,
Bcell_vs_Tcell_For_antiBLTA =
anti_BTLA.Purified_B_cell-anti_BTLA.Purified_CD4_T_cell,
levels = design )
6.5 差异分析
1️⃣ 现在我们比较一下在B细胞中anti-BLTA组和Control组之间的差异表达基因。
fit2 <- contrasts.fit(fit, cm)
fit2 <- eBayes(fit2)
res_antiBLTA_vs_Control_For_Bcell <- topTable(fit2,
coef = "antiBLTA_vs_Control_For_Bcell",
n = Inf,
#p.value = 0.05
)

2️⃣ 再比较一下在T细胞中anti-BLTA组和Control组之间的差异表达基因。
res_antiBLTA_vs_Control_For_Tcell <- topTable(fit2,
coef = "antiBLTA_vs_Control_For_Tcell",
n = Inf,
#p.value = 0.05
)

3️⃣ 再比较一下Control组内T细胞和B细胞之间的差异表达基因。
res_Bcell_vs_Tcell_For_Control <- topTable(fit2,
coef = "Bcell_vs_Tcell_For_Control",
n = Inf,
#p.value = 0.05
)
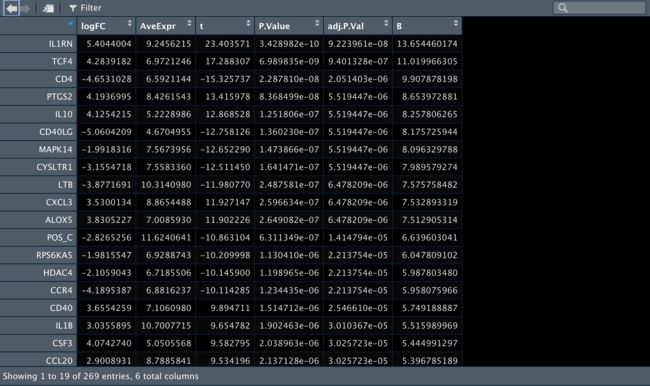
4️⃣ 再比较一下anti-BLTA组内T细胞和B细胞之间的差异表达基因。
res_Bcell_vs_Tcell_For_antiBLTA <- topTable(fit2,
coef = "Bcell_vs_Tcell_For_antiBLTA",
n = Inf,
#p.value = 0.05
)

7可视化
我们直接用火山图可视化对比一下吧。 这里我们把阈值设置为|logFC|>1,pvalue<0.05。
library(EnhancedVolcano)
library(patchwork)
p1 <- EnhancedVolcano(res_antiBLTA_vs_Control_For_Bcell,
lab = rownames(res_antiBLTA_vs_Control_For_Bcell),
x = 'logFC',
y = 'P.Value',
title = 'B cell (anti-BTLA vs Control)',
pointSize = 3.0,
labSize = 6.0,
legendPosition = 'right',
pCutoff = 0.05,
FCcutoff = 1)
p2 <- EnhancedVolcano(res_antiBLTA_vs_Control_For_Tcell,
lab = rownames(res_antiBLTA_vs_Control_For_Tcell),
x = 'logFC',
y = 'P.Value',
title = 'T cell (anti-BTLA vs Control)',
pointSize = 3.0,
labSize = 6.0,
legendPosition = 'right',
pCutoff = 0.05,
FCcutoff = 1)
p3 <- EnhancedVolcano(res_Bcell_vs_Tcell_For_Control,
lab = rownames(res_Bcell_vs_Tcell_For_Control),
x = 'logFC',
y = 'P.Value',
title = 'B cell vs T cell (Control)',
pointSize = 3.0,
labSize = 6.0,
legendPosition = 'right',
pCutoff = 0.05,
FCcutoff = 1)
p4 <- EnhancedVolcano(res_Bcell_vs_Tcell_For_antiBLTA,
lab = rownames(res_Bcell_vs_Tcell_For_antiBLTA),
x = 'logFC',
y = 'P.Value',
title = 'B cell vs T cell (anti-BLTA)',
pointSize = 3.0,
labSize = 6.0,
legendPosition = 'right',
pCutoff = 0.05,
FCcutoff = 1)
wrap_plots(p1,p2,p3,p4)

点个在看吧各位~ ✐.ɴɪᴄᴇ ᴅᴀʏ 〰

本文由 mdnice 多平台发布