医学图像配准:A Rigid Registration Method in TEVAR
A Rigid Registration Method in TEVAR
TEVAR 中的一种刚性配准方法
摘要
Since the mapping relationship between definitized intra-interventional 2D X-ray and undefined pre-interventional 3D Computed Tomography(CT) is uncertain, auxiliary positioning devices or body markers, such as medical implants, are commonly used to determine this relationship. However, such approaches can not be widely used in clinical due to the complex realities. To determine the mapping relationship, and achieve a initializtion post estimation of human body without auxiliary equipment or markers, proposed method applies image segmentation and deep feature matching to directly match the 2D X-ray and 3D CT images. As a result, the well-trained network can directly predict the spatial correspondence between arbitrary 2D X-ray and 3D CT. The experimental results show that when combining our approach with the conventional approach, the achieved accuracy and speed can meet the basic clinical intervention needs, and it provides a new direction for intra-interventional registration.
由于确定的术中(intra-interventional) 2D X 射线和未知的术前(pre-interventional ) 3D 断层扫描 (CT) 之间的映射关系是不确定的,因此通常使用辅助定位设备或身体标记点(markers),例如医疗植入物来确定这种关系。 然而,由于复杂性,这种方法不能广泛应用于临床。 为了确定映射关系,并在没有辅助设备的情况下实现人体姿态估计,该方法应用图像分割和深度特征匹配来直接配准2D X射线和3D CT图像。 因此,训练有素的网络可以直接预测任意 2D X 射线和 3D CT 之间的空间对应关系。 实验结果表明,当我们的方法与传统方法相结合时,所达到的准确性和速度可以满足基本的临床干预需求,为介入内配准提供了新的方向。
介绍
Thoracic Endovascular Aortic Repair (TEVAR) is the preferred treatment for thoracic aortic dilatation diseases as it allows the low mortality and complications [11]. Interventional therapy is often performed on two-dimensional(2D) medical images such as X-ray. Due to the lack of clear development of vessels under X-ray fluoroscopy, contrast agents which often cause serious
胸主动脉血管内修复术 (TEVAR) 是胸主动脉扩张疾病的首选治疗方法,因为它具有较低的死亡率和并发症 [11]。 治疗通常在二维 (2D) 医学图像上执行,例如 X 射线。 由于 X 线透视下血管发育不清晰,经常导致严重的造影剂
complications have to be used in interventional therapy. This paper presents a medical image registration approach based on transformer to accurately develop the aorta and its branches directly on X-ray fluoroscopy. Due to the position of aortic arch is fixed by the arterial ligament and three arterial branches, it is considered immobile in proposed approach. The feasibility of this idea will be validated in Section.4.2.
并发症必须用于介入治疗。 本文提出了一种基于变压器的医学图像配准方法,可直接在 X 射线中准确地显示主动脉及其分支。 由于主动脉弓的位置由动脉韧带和三个动脉分支固定,因此在建议的方法中被认为是固定的。 这个想法的可行性将在第 4.2 节中得到验证。
Machine learning-based medical image registration methods are mostly divided into two cat- egories: optimization-based and learning-based approaches. Optimization-based conventional approaches project the 3D CT data to 2D virtual X-ray, according to a series of projection param- eters. Then searching for the most similar image to the real X-ray iterativly and determining its spatial transformation parameters.which is difficult to meet the requirement of real-time registration suffer from extremely high computational cost and sensition of initial estimates[7]. Although the recent development of deep learning have been successfully applied to various medical applications [9, 12–14], such techniques have been barely used for 3D CT data to 2D virtual X-ray image registra- tion. Even some works simply applied Convolutional Neural Networks (CNNs), autoencoder (AEs) or other network structures to conduct feature extraction[17, 21]or similarity measurement [1, 3], which direct predict the registration parameter, they haven’t solve the problem of time-consuming of iteratively optimization. More importantly, considering the certainty of intra-interventional human position and the uncertainty of pre-interventional CT data acquisition position, researchers had to choose positioning from two perspectives[5, 8] or using traumatic markers like medical implants[8] to solve the problems. In other words, these methods are not widely used in clinic.
基于机器学习的医学图像配准方法主要分为两类:基于优化的方法和基于学习的方法。根据一系列投影参数,基于优化的传统方法将 3D CT 数据投影到 2D 虚拟 X 射线。然后迭代寻找与真实X射线最相似的图像并确定其空间变换参数,难以满足实时配准的要求,计算成本极高,初始估计敏感[7]。 尽管最近深度学习的发展已成功应用于各种医学应用 [9, 12-14],但此类技术几乎没有用于 3D CT 数据到 2D 虚拟 X 射线图像配准。甚至有些作品只是简单地应用卷积神经网络(CNNs)、自动编码器(AEs)或其他网络结构来进行特征提取[17, 21]或相似度测量[1, 3],直接预测配准参数,他们也没有解决迭代优化耗时的问题。更重要的是,考虑到术中人体位置姿态的确定性和术前CT数据采集位置的不确定性,研究人员不得不从两个角度选择定位[5, 8]或使用医疗植入物等创伤标记物[8]来解决问题。 换言之,这些方法在临床上并未广泛使用。
Since pre-interventional 3D CT can provide accurate anatomical information, this paper adopts 2D-3D image registration method, The aim is to establish a direct connection between 2D X-ray and 3D CT. The main ideas of proposed method are as follows: two segmentation networks been used to segment skeletal features from two dimension. Then the extracted features are put into the crossmodal matching transformer module to matching features of different dimensions. After the process of learning mapping relationships, we embed five positioning parameters available in interventional imaging equipment into our network to improved the ability of the network. The main contributions of our research are:
由于术前3D CT可以提供准确的解剖信息,本文采用2D-3D图像配准方法,目的是建立2D X射线和3D CT之间的直接连接。 所提出方法的主要思想如下:使用两个分割网络从二维分割骨骼特征。 然后将提取的特征放入交叉模态匹配变换器模块中,以匹配不同维度的特征。 在学习映射关系的过程之后,我们将术中成像设备中可用的五个定位参数嵌入到我们的网络中,以提高网络的能力。 我们研究的主要贡献是:
• Application of medical image registration technology in the field of TEVAR interventional therapy.
• The proposed approach can directly match 2D and 3D images whose latent features. This provide a solution for the precise localization in TEVAR under X-ray, and can potentially reduce the use of contrast agents and the exposure time under radiation.
• 医学影像配准技术在TEVAR介入治疗领域的应用。
• 所提出的方法可以直接匹配具有潜在特征的 2D 和 3D 图像。 这为 X 射线下 TEVAR 中的精确定位提供了解决方案,并可能减少造影剂的使用和辐射下的暴露时间。
相关研究
2.1 基于学习的医学图像分割
Although image segmentation technology has been widely used in the field of medicine, the methods used in clinic are still limited. Medical images have abundant spatial information (such as complex texture structure), and the process of network downsampling is easy to lose these spatial information. Encoder-decoder networks with symmetric structures[10, 12]. can be better preserve these spatial information. Using the same method, Cicek et al. replaced the two-dimensional convolution layer with the three-dimensional convolution layer to construct 3D U-Net, and realized the end-to-end processing of 3D CT[2]. Although deep learning have shown significant progress compared with conventional algorithms, the process of medical image tagging is time-consuming and labor-consuming, which limits the further development of deep learning algorithms in clinic. Researchers consider more diverse ways of using unlabeled data[18, 20], but such segmentation methods are not suitable for registration tasks that are extremely sensitive to spatial position.
虽然图像分割技术在医学领域得到了广泛的应用,但在临床上使用的方法仍然有限。医学图像具有丰富的空间信息(如复杂的纹理结构),网络下采样的过程很容易丢失这些空间信息。具有对称结构的编码器-解码器网络[10, 12]。可以更好地保存这些空间信息。 Cicek 等人使用相同的方法。用三维卷积层代替二维卷积层构建3D U-Net,实现了3D CT的端到端处理[2]。尽管深度学习与传统算法相比取得了显着进步,但医学图像标注的过程费时费力,限制了深度学习算法在临床上的进一步发展。研究人员考虑了更多样化的使用未标记数据的方法[18, 20],但这种分割方法不适用于对空间位置极为敏感的配准任务。
2.2 多模态数据融合
It is a natural idea to fuse the same features extracted from two- and three-dimensional neural networks. S-PCNN[19] divides the source image into several blocks, then calculate the spatial frequency (SF) of the blocks as linking strength beta of the PCNN. It is used to extract the medical imageures find the best oscillation frequency graph (OFG) iteratively. Although combined the infor- mation of multi-modal images, the time-consuming of divided blocks and iteration is not allowed in real-time medical image registration. Parallel cross CNN (PCCNN) model[15] extracts two group features of CNN in parallel through a couple of deep CNN data transform flows. The information of two streams are fused together after the first fully connected layers in each stream. After the final Softmax regression, the 1024-dimensional image feature vector is classified and recognized. The method which introduced depthwise separable convolution combined with depthwise and point- wise convolution to replace the standard convolution as a basic convolution module[4], respectively applies the convolution in different channels, and applies a 1 × 1 convolution to combine separate features generated by the depthwise convolution to reduce the complexity and computational cost of model. The method of fussing information of unaligned multimodal language sequences gives us enlightenment[16]. By the method of multimodal transformer, they address the above issues in an end-to-end manner without explicitly aligning the audio, language, vision.
融合从二维和三维神经网络中提取的相同特征是很自然的想法。 S-PCNN[19] 将源图像分成若干块,然后计算块的空间频率(SF)作为 PCNN 的链接强度 beta。它用于提取医学图像,以迭代方式找到最佳振荡频率图(OFG)。虽然结合了多模态图像的信息,但在实时医学图像配准中不允许分块和迭代的耗时。并行交叉CNN(PCCNN)模型[15]通过几个深度CNN数据变换流并行提取CNN的两组特征。两个流的信息在每个流中的第一个全连接层之后融合在一起。经过最后的Softmax回归,对1024维的图像特征向量进行分类识别。
引入depthwise separable convolution结合depthwise和point-wise卷积来代替标准卷积作为基本卷积模块的方法[4],分别在不同的通道上应用卷积,并应用一个1×1的卷积来组合由深度卷积以降低模型的复杂性和计算成本。对未对齐的多模态语言序列进行信息处理的方法给了我们启示[16]。通过多模态转换器的方法,他们以端到端的方式解决上述问题,而无需明确对齐音频、语言、视觉。
方法论
3.1 特征提取
Our approach starts with extracting skeletons from 2D X-ray and 3D CT images. In particular, we train two UNet models for 2D skeleton segmentation and 3D skeleton segmentation tasks, as UNet has been successfully applied in various image segmentation tasks [2, 12]. After they are well trained, their encoders can consequently provide strong skeleton-related representations, and thus we take them as the input for downstream tasks.
我们的方法从从 2D X 射线和 3D CT 图像中提取骨骼开始。 特别是,我们为 2D 分割和 3D 分割任务训练了两个 UNet 模型,因为 UNet 已成功应用于各种图像分割任务 [2, 12]。 在他们训练有素之后,他们的编码器可以提供强大的相关表示,因此我们将它们作为下游任务的输入。
The 2D encoder is made up of five blocks that contains three 2D convolution units( a 3 × 3 convolution layer, a batch normalization layer (BN) and a ReLU activation) and a 3 × 3 max pooling layer with stride of two. Meanwhile, the 3D encoder is also made up of five blocks, where the first block has two 3D convolution units ( a 3 × 3 × 3 convolution layer, a batch normalization layer (BN) and a ReLU activation ) and a 3 × 3 × 3 max pooling with stride of two while Each of the rest four blocks containing three 3D convolution units and a 3D max pooling. In this paper, the output of the 2D and 3D encoder are 512-channel 2D feature maps and 64-channel 3D feature maps, representing the skeleton locations and appearance information of the 2D and 3D input.
-
2D 编码器由五个块组成,其中包含三个 2D 卷积单元(一个 3 × 3 卷积层、一个批量归一化层 (BN) 和一个 ReLU 激活)和一个 3 × 3 最大池化层,步长为 2。
-
同时,3D 编码器也由五个块组成,其中第一个块有两个 3D 卷积单元(一个 3 × 3 × 3 卷积层、一个批归一化层 (BN) 和一个
ReLU 激活)和一个 3 × 3 × 3 个步长为 2 的最大池化,而其余四个块中的每一个都包含三个 3D 卷积单元和一个 3D
最大池化。 本文中2D和3D编码器的输出是512通道2D特征图和64通道3D特征图,分别表示2D和3D输入的骨架位置和外观信息。
3.2 深度特征匹配
After the feature extraction of 2D and 3D encoders way, the 2D and 3D skeleton representations are directly combined for registration rather than previous approaches [6, 8] which convert 3D image to 2D then registration.
在 2D 和 3D 编码器的特征提取方式之后,2D 和 3D 骨骼表示直接组合进行配准,而不是以前的方法 [6, 8] 将 3D 图像转换为 2D 然后配准。
Full network structure and more details please see the official article published in August 2021.
完整的网络结构和更多详细信息请参见2021年8月发布的官方文章。
3.3 附加参数
Five known parameters: the angle of L-arm rotation(α), the angle of pivot rotation(β), the angle of Carm rotation(γ), the distance from source to patient(SSD), the distance from source to detector(SI D), and the coordinate of isocenter in pre-interventional CT data.
五个已知参数:L臂旋转角度(α)、枢轴旋转角度(β)、卡姆旋转角度(γ)、源到患者的距离(SSD)、源到探测器的距离( SI D),以及介入前 CT 数据中等中心的坐标。

实验结果与分析
4.1 实现细节
4.1.1 Dataset. Pre-interventional CT images of 1162 patients suffering thoracic aortic aneurysm or B aortic dissection were collected for training of 3D segmentation network. Among them, 1000 were used for training and 162 for validation. A total of 49176 frames of X-ray examination parameters were counted in 162 patients. Their distribution is shown in Fig.3. According to the distribution range of the above parameters and random sampling of the position coordinates of isocenter in CT space, the clinical projection transformation space is constructed.
收集了 1162 名胸主动脉瘤或 B 主动脉夹层患者的术前 CT 图像,用于 3D 分割网络的训练。 其中,1000 个用于训练,162 个用于验证。 162例患者共统计49176帧X线检查参数。 它们的分布如图3所示。 根据上述参数的分布范围和CT空间等中心位置坐标的随机采样,构建临床投影变换空间。
Virtual intra-interventional X-rays are generated by ray casting to CT data with known clinical spatial transformation parameters. The input to 2D segmentation network is a 352 × 512 image with 1 channels, which is cut out from the upper part of virtual X-ray for removing the part of the diaphragm with larger motion. The corresponding skeleton label is segmented from CT data using the threshold method. After that, the 873 groups pre-interventional CT images are tracing to generate 87300 virtual X-rays according to the range shown in table.1 and distribution shown in Fig.3. where d represents the scanning diameter of CT data.
通过将射线投射到具有已知临床空间转换参数的 CT 数据来生成虚拟介入内 X 射线。 2D 分割网络的输入是一个 352 × 512 的 1 通道图像,它是从虚拟 X 射线的上部切出的,用于去除振动较大的部分。 相应的骨架标签是使用阈值方法从 CT 数据中分割出来的。 之后,873组介入前CT图像按照表1所示的范围和图3所示的分布进行追踪,生成87300张虚拟X射线。 其中 d 表示 CT 数据的扫描直径。


4.1.2 Training details. We implement the proposed approach under the Pytorch framework with GPU acceleration. 3D segmentation network was trained by 1000 sets of CT data and their skeletal labels and verified by 162 sets. And 2D skeleton segmentation network was trained by 543 groups virtual X-ray with a total of 543000 images and validated by 162 groups with a total of 97000 images. These virtual X-ray is generated according to the parameter distribution of clinical X-ray, as described in Section.4.1.1.
4.1.2 培训细节。 我们在具有 GPU 加速的 Pytorch 框架下实现了所提出的方法。 3D 分割网络由 1000 组 CT 数据及其骨骼标签训练,并由 162 组验证。 2D骨骼分割网络由543组虚拟X射线共543000张图像进行训练,并由162组共97000张图像进行验证。 这些虚拟 X 射线是根据临床 X 射线的参数分布生成的,如第 4.1.1 节所述。
Full network structure and more details please see the official article published in August 2021.
完整的网络结构和更多详细信息请参见2021年8月发布的官方文章。
4.2 主动脉弓刚性配准可行性分析
During interventional surgery, the doctor estimated the location of the branches of the aorta according to the position of the bone and the intraoperative guide wire under X-ray. In order to verify the rigid registration of bone can locate the aortic arch and its branches, we measured the displacement of vertebral bone, rib, guide wire and proximal stent in 309 groups intra-interventional X-ray. Because the vessel can not be clearly developed under X-ray, we use the displacement of the proximal stent post-intenventional to approximate estimates the displacement of the aortic arch and its branches. As a validation of the feasibility of our method.
介入手术时,医生根据骨的位置和术中导丝在X线下估计主动脉分支的位置。 为了验证骨刚性配准能够定位主动脉弓及其分支,我们测量了309组介入X线片中椎骨、肋骨、导丝和近端支架的位移。 由于血管在 X 线下不能清楚地显影,我们使用近端支架的位移来近似估计主动脉弓及其分支的位移。 作为对我们方法可行性的验证。
Since the average dynamic displacement error of branches vessels on aortic arch is about 2.95mm, the opening diameter of the left subclavian artery is about 10 mm and the respect of proximal Table 2. The displacement of intra-interventional vertebral bone, rib, guide wire and post-interventional proximal stent
由于主动脉弓上分支血管的平均动态位移误差约为2.95mm,左锁骨下动脉开口直径约为10mm,近端方面见表2 介入椎骨、肋骨、导丝的位移 和介入后近端支架
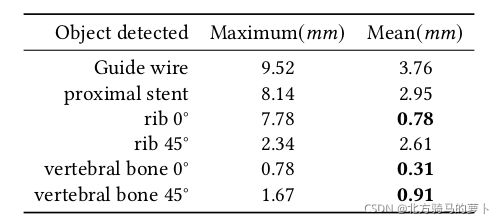
anchoring area over 15 mm, we have good reason to think that rigid registration can achieve the initial localization of the aorta in TEVAR.
锚定区域超过 15 mm,我们有充分的理由认为刚性配准可以在 TEVAR 中实现主动脉的初始定位。
4.3 注册实验
We compare our approach to a learning-based(CNN[8]) and two optimization-based methods. Cross-correlation(Opt-CC) and normalized mutual information(Opt-NMI) are used to measure the similarity respectively and Powell algorithm is used to optimize similarity iterativly. In addition, we compare the performance of the optimization-based aprroach using two learning-based aprroaches as initializers(Denoted as CNN+Opt and Cross-Transformer+Opt).
我们将我们的方法与基于学习的(CNN[8])和两种基于优化的方法进行比较。 分别使用互相关(Opt-CC)和归一化互信息(Opt-NMI)来衡量相似度,并使用鲍威尔算法迭代优化相似度。 此外,我们比较了使用两种基于学习的 aprroaches 作为初始化器(表示为 CNN+Opt 和 Cross-Transformer+Opt)的基于优化的 aprroach 的性能。
The position of the aortic branch on DSA image at same time is taken as the gold standard, and the registration error of these approaches(mean target registration error, mTRE) is measured. The gold standard data is untrained. The gross failure rate(GFR) is defined as the percentage of th tested cases with a TRE greater than 10mm. And the average time cost of registration(Reg.time) and out-plane rotation error(Rot.error) additionally demonstrate of the superiority of the approach. For detailed details of the genuine articles published in August.
同时以主动脉分支在DSA图像上的位置为金标准,测量这些方法的配准误差(平均目标配准误差,mTRE)。 黄金标准数据未经训练。 总失效率(GFR)定义为 TRE 大于 10mm 的测试案例的百分比。 配准的平均时间成本(Reg.time)和平面外旋转误差(Rot.error)进一步证明了该方法的优越性。 详细了解八月份发表的正版文章。
Experimental results show that the accuracy and speed of registration are improved to some extent by using learning-based approach to initialize the optimization-based approach. The average displacement error of 4.7mm is considered to be of clinical significance in TEVAR operation. Although this method is still not very robust in the comprehensive performance of registration, it is carried out without additional positioning equipment or markers.
Experimental results show that the accuracy and speed of registration are improved to some extent by using learning-based approach to initialize the optimization-based approach. The average displacement error of 4.7mm is considered to be of clinical significance in TEVAR operation. Although this method is still not very robust in the comprehensive performance of registration, it is carried out without additional positioning equipment or markers.
结论
In this paper, we propose a novel 3D-2D medical image registration approach. The main novelty of this paper is the proposed feature transform method that allows the generated feature maps retain the initial pose estimation of any 3D CT and corresponding 2D X-ray, without any other positioning items. The excellent experimental results shows that the proposed approach is suitable for basic clinical intervention usage and provides a new direction for intra-interventional registration.
在本文中,我们提出了一种新颖的 3D-2D 医学图像配准方法。 本文的主要新颖之处在于所提出的特征变换方法,该方法允许生成的特征图保留任何 3D CT 和相应 2D X 射线的初始姿态估计,而无需任何其他定位项。 优异的实验结果表明,所提出的方法适用于基本的临床干预使用,并为介入内配准提供了新的方向。
Although deep learning has been widely used in the field of computer vision, its imprecision limits its application in the medical image registration. The approach of directly regression parameters is constrained by network performance, whose performance is not accurate enough to classify parameters. Pixel level accuracy is the most important problem to be solved in the field of registration. Other areas, such as image recognition, require translation rotation invariance, which is strictly prohibited in medical image registration. Therefore, the reference network structure should be more rigorous.
尽管深度学习在计算机视觉领域得到了广泛的应用,但其不精确性限制了其在医学图像配准中的应用。 直接回归参数的方法受到网络性能的限制,其性能不足以准确地对参数进行分类。 像素级精度是配准领域最需要解决的问题。 其他领域,如图像识别,需要平移旋转不变性,这在医学图像配准中是严格禁止的。 因此,参考网络结构应该更加严谨。
