Python使用支持向量机(SVM)方法对UCI 乳腺癌诊断数据集二分类任务
数据集:本文数据来自UCI repository美国威斯康星州的乳腺癌诊断数据集,实验所用的编译环境为python3.6,主要引用numpy、sklearn、pandas、matplotlib等库。
涉及到的方法:机器学习SVM,混淆矩阵,选择核函数,调参
加载数据集
# 加载数据
path = 'D:/code/breast_cancer/wdbc.data'
names = ['ID number', 'Diagnosis',
'M_radius', 'M_texture', 'M_perimeter', 'M_area', 'M_smoothness', 'M_compactness', 'M_concavity',
'M_concave points', 'M_symmetry', 'M_fractal dimension',
'SE_radius', 'SE_texture', 'SE_perimeter', 'SE_area', 'SE_smoothness', 'SE_compactness', 'SE_concavity',
'SE_concave points', 'SE_symmetry', 'SE_fractal dimension',
'W_radius', 'W_texture', 'W_perimeter', 'W_area', 'W_smoothness', 'W_compactness', 'W_concavity',
'W_concave points', 'W_symmetry', 'W_fractal dimension']
data= pd.read_csv(path, header=None, names=names)
#data.to_csv('D:/code/breast_cancer/wdbc.csv')
# 显示数据
pd.set_option('display.max_columns', None)
#print(data.columns)
#print(data.head())
#print(data.describe())
# 诊断结果B良性:0,M恶性:1
#print(type(data))
data['Diagnosis'] = data['Diagnosis'].map({'M':1, 'B':0})
#print(data)
预处理
为了减少计算量,我们可以对数据进行降维处理,用少量的特征代表数据的特性,增强模型的泛化能力,避免数据过拟合。由于三种计算值表示的特征相同,因此这里仅选择均值进行描述。
# 特征字段分组,mean、se、worst
feature_mean = list(data.columns[2:12])
feature_se = list(data.columns[12:22])
feature_worst = list(data.columns[22:32])
# 数据清洗,去掉ID
#data.drop(columns=['ID number'],axis=1,inplace=True)
# 诊断结果可视化
sns.countplot(data['Diagnosis'],label='Count')
corr = data[feature_mean].corr()
plt.figure(figsize=(14, 14))
sns.heatmap(corr, annot=True)
plt.show()
通过热力图看出,半径、周长和区域相关度,紧密度、凹面和凹点的相关度都很高,因此各选择一个特征作为代表。最终选用平均半径、平均纹理、平均平滑度、平均紧密度、平均对称性和平均分形维数6种特征进行描述。
# 缩减属性(去除高度相关的,减小计算量)
feature_remain = ['M_radius', 'M_texture', 'M_smoothness',
'M_compactness', 'M_symmetry', 'M_fractal dimension']
将数据随机混合并按照6:2:2的比例分割为训练集(341例)、验证集(114例)和测试集(114例)。再采用 Z-Score 标准化数据,保证每个特征维度的数据均值为0,方差为1。
# 抽取数据,6:2:2
train, test1 = train_test_split(data, test_size=0.4, random_state=111)
val, test = train_test_split(test1, test_size=0.5, random_state=121)
# print(len(train), len(val), len(test))
train_x = train[feature_remain]
train_y = train['Diagnosis']
val_x = val[feature_remain]
val_y = val['Diagnosis']
test_x = test[feature_remain]
test_y = test['Diagnosis']
# 对数据进行z-score归一化
ss = StandardScaler()
train_x = ss.fit_transform(train_x)
val_x = ss.fit_transform(val_x)
test_x = ss.fit_transform(test_x)
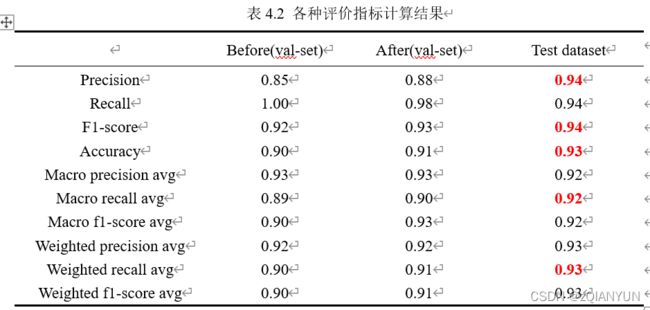
评估指标说明
- 精确度: 精确度直观表示了分类器不将负样本标记为正样本的能力。 Precision=TP/(TP+FP)×100%
- 召回率:召回率表示了分类器找到所有正样本的能力。 Recall=TP/(TP+FN)×100%
- F1-score:F1分数是精确率和召回率的调和平均数,最大为1,最小为0。 F1-score=
(2Precision×Recall)/(Precision+Recall) - 支持度:每个类中正确的目标值。
- 准确率:代表分类器对整个样本判断正确的比重。
Accuracy=(TP+TN)/(TP+FP+FN+TN)×100% - 宏观平均值:先针对每个类计算评估指标,再对他们取平均。 Macro Precesion=1/n ∑_(i=1)^n▒P_i
Macro Recall=1/n ∑_(i=1)^n▒R_i Macro F1=1/n ∑_(i=1)^n▒F_i - 加权平均值:根据每个类的样本量,给每个类赋予不同的权重,再取平均值。
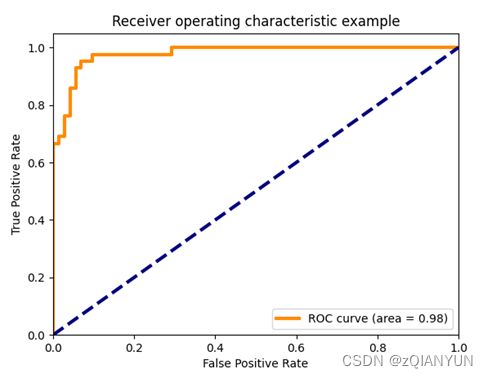
- ROC曲线:可评判分类结果的好坏。横坐标为FPR(假正率),计算分类器错认为正类的负样本的比例。纵坐标为TPR(真正率),代表分类器所识别的正样本所占所有正样本的比例。ROC曲线下方的面积为AUC,其值越大说明模型的性能越好。
FPR=FP/(FP+TN) TPR=TP/(TP+FN)
核函数选择和训练
SVM模型中的参数包括:C, kernel, degree, gamma, coef0, shrinking, probability, tol, cache_size, class_weight, verbose, max_iter, decision_function_shape, random_state,其中c和gamma最为重要。c是惩罚系数,即对误差的宽容度。c越高,说明越不能容忍出现误差,容易过拟合;c越小,越容易欠拟合。而gamma隐含地决定了数据映射到新的特征空间后的分布,gamma越大,支持向量越少,gamma值越小,支持向量越多。
为了筛选出合适的kernel,我们在验证集上计算了每种核函数使用默认参数时的准确率进行了对比,结果中高斯核的表现最佳,准确率达到0.904。
# 创建SVM分类器
print("-------调参前---------")
clf = svm.SVC()
clf.fit(train_x, train_y)
predictions = clf.predict(val_x)
print(confusion_matrix(val_y, predictions))
print(classification_report(val_y, predictions))
# Kernel = ["linear","poly","rbf","sigmoid"]
# print("-------选择核函数---------")
#
# for kernel in Kernel:
# model= svm.SVC(kernel = kernel).fit(train_x,train_y)
# validation = model.predict(val_x)
# print(kernel,'val acc: ', metrics.accuracy_score(validation, val_y))
因此SVM模型使用高斯核,并在验证集上通过网格选择法筛选出最优的c和gamma的组合。这里将c的参数设置为[0.1, 1, 10, 100, 1000],gamma的参数设置为[1, 0.1, 0.01, 0.001, 0.0001],使用网格搜索函数GridSearchCV( )进行筛选,为25个候选对象各拟合5次,共计125次拟合。最后选出得分最佳的组合为c=100,gamma=0.01。
model = svm.SVC(kernel="linear")
model.fit(train_x, train_y)
# 验证集
predictions = model.predict(val_x)
print(confusion_matrix(val_y, predictions))
print(classification_report(val_y, predictions))
plot_confusion_matrix(model, val_x, val_y, values_format='d', display_labels=['malignant', 'benign'])
plt.show()
# param_grid = {'C':[0.1,1,10,100,1000], 'gamma':[1,0.1,0.01,0.001,0.0001]}
param_grid = {'C':[0.1,1,5,10,15,100,1000]}
grid_model = GridSearchCV(svm.SVC(), param_grid, verbose=3)
grid_model.fit(train_x, train_y)
#print(grid_model)
print(grid_model.best_params_)
grid_predictions_val = grid_model.predict(val_x)
print(confusion_matrix(val_y, grid_predictions_val))
print(classification_report(val_y, grid_predictions_val))
plot_confusion_matrix(grid_model, val_x, val_y, values_format='d', display_labels=['malignant', 'benign'])
plt.show()
用调整后的网络进行测试,得出最终结果
# 测试集
grid_predictions = grid_model.predict(test_x)
print(confusion_matrix(test_y, grid_predictions))
print(classification_report(test_y, grid_predictions))
# plot_confusion_matrix(grid_model, test_x, test_y, values_format='d', display_labels=['malignant', 'benign'])
# plt.show()