mpf6_Time Series Data_quandl_更正kernel PCA_AIC_BIC_trend_log_return_seasonal_decompose_sARIMAx_ADFull
In financial portfolios, the returns on their constituent(/kənˈstɪtʃuənt/组成的,构成的) assets depend on a number of factors, such as macroeconomic and microeconomical conditions, and various financial variables. As the number of factors increases, so does the complexity involved in modeling portfolio behavior. Given that computing resources are finite, coupled with time constraints, performing an extra computation for a new factor only increases the bottleneck on portfolio modeling calculations. A linear technique for dimensionality reduction is Principal Component Analysis (PCA). As its name suggests, PCA breaks down the movement of portfolio asset prices into its principal components, or common factors, for further statistical analysis. Common factors that don't explain much of the movement of the portfolio assets receive less weighting in their factors and are usually ignored. By keeping the most useful factors, portfolio analysis can be greatly simplified without compromising on computational time and space costs.
In statistical analysis of time series data, it is important for the data to be stationary in order to avoid spurious(/ˈspjʊriəs/虚假的,伪造的) regression. Non-stationary data may be generated by an underlying process that is affected by a trend, a seasonal effect, presence of a unit root, or a combination of all three. The statistical properties of non-stationary data, such as mean and variance, changes over time. Non-stationary data needs to be transformed into stationary data for statistical analysis to produce consistent and reliable results. This can be achieved by removing the trend and seasonality components. Stationary data can there after be used for prediction or forecasting.
In this chapter, we will cover the following topics:
- Performing PCA on the Dow and its 30 components
- Reconstructing the Dow index
- Understanding the difference between stationary and non-stationary data
- Checking data for stationarity
- Types of stationary and non-stationary processes
- Using the Augmented Dickey-Fuller Test to test the presence of a unit root
- Making stationary data by detrending, differencing, and seasonal decomposing
- Using an Autoregressive Integrated Moving Average for time series prediction and forecasting
The Dow Jones industrial average and its 30 components
The Dow Jones Industrial Average (DJIA) is a stock market index that comprises the 30 largest US companies. Commonly known as the Dow, it is owned by S&P Dow Jones Indices LLC and computed on a price-weighted basis (see https://www.spglobal.com/spdji/en/index-family/equity/us-equity/dow-jones-averages/#overview for more information on the Dow).
Downloading Dow component datasets from Quandl
The following code retrieves the Dow component datasets from Quandl. The data provider that we will be using is WIKI Prices, a community formed by members of the public and that provides datasets free of charge back to the public. Such data isn't free from errors, so please use them with caution. At the time of writing, this data feed is no longer supported actively by the Quandl community, though past datasets are still available for use. We will download historical daily closing prices for 2017:
https://blog.csdn.net/Linli522362242/article/details/93617948
https://blog.csdn.net/Linli522362242/article/details/121172551
!pip3 install quandl
https://blog.csdn.net/Linli522362242/article/details/121172551

The wiki_symbols variable contains a list of Quandl codes that we use for downloading. Notice that in the parameter arguments of quandl.get() , we specified column_index=11. This tells Quandl to download only the 11th column of each dataset, which coincides with the adjusted daily closing prices. The datasets are downloaded into our df_components variable as a single pandas DataFrame object.
import quandl
QUANDL_API_KEY = 'sKqHwnHr8rNWK-3s5imS'
quandl.ApiConfig.api_key = QUANDL_API_KEY
SYMBOLS = [
'AAPL','MMM', 'AXP', 'BA', 'CAT',
'CVX', 'CSCO', 'KO', 'DD', 'XOM',
'GS', 'HD', 'IBM', 'INTC', 'JNJ',
'JPM', 'MCD', 'MRK', 'MSFT', 'NKE',
'PFE', 'PG', 'UNH', 'UTX', 'TRV',
'VZ', 'V', 'WMT', 'WBA', 'DIS',
]
wiki_symbols = ['WIKI/%s' % symbol
for symbol in SYMBOLS
]
# https://docs.data.nasdaq.com/docs/parameters-2
# column_index
# Request a specific column. Column 0 is the date column
# and is always returned. Data begins at column 1.
df_components = quandl.get( wiki_symbols,
start_date='2017-01-01',
end_date='2017-12-31',
column_index=11 # Adj.close
)
df_components.columns = SYMBOLS # Renaming the columnsprint(df_components)def get_quandl_dataset( api_key, code, start_date, end_date, column_index ):
"""Obtain and parse a quandl dataset in Pandas DataFrame format
Quandl returns dataset in JSON format, where data is stored as a
list of lists in response['dataset']['data'], and column headers
stored in response['dataset']['column_names'].
Args:
api_key: Quandl API key
code: Quandl dataset code
Returns:
df: Pandas DataFrame of a Quandl dataset
"""
# https://docs.data.nasdaq.com/docs/in-depth-usage
# https://data.nasdaq.com/api/v3/datasets/{database_code}/{dataset_code}.json?api_key=sKqHwnHr8rNWK-3s5imS
# https://docs.data.nasdaq.com/docs/parameters-2
# example
# https://data.nasdaq.com/api/v3/datasets/wiki/AAPL.json?api_key=sKqHwnHr8rNWK-3s5imS
# &start_date=2017-01-01
# &end_date=2017-06-30
base_url = "https://data.nasdaq.com/api/v3/datasets/"
url_suffix = ".json?api_key="
para = "&start_date={}&end_date={}&column_index={}".format( start_date, end_date, column_index )
# Fetch the JSON response
u = urlopen(base_url + code + url_suffix + api_key + para)
# https://data.nasdaq.com/api/v3/datasets/WIKI/AAPL.json?api_key=sKqHwnHr8rNWK-3s5imS&start_date=2017-01-01&end_date=2017-06-30
response = json.loads( u.read().decode('utf-8') )
# Format the response as Pandas Dataframe
df = pd.DataFrame( response['dataset']['data'],
columns = ['Date', 'Adj.Close']
)
return df
# Input your own API key here
api_key = "sKqHwnHr8rNWK-3s5imS" #"gwguNnzq_4xR18V7ChED"
# Quandl code for six US companies
# {database_code}/{dataset_code}
start_date = "2017-01-01"
end_date = "2017-12-31"
column_name = 'Adj.Close'
dfs = []
# Get the DataFrame that contains the WIKI data for each company
for code in wiki_symbols:
df = get_quandl_dataset( api_key, code, start_date, end_date, 11 )
df.rename( columns={ 'Adj.Close': code[5:] },
# WIKI/AAPL ==> AAPL
inplace=True )
# or df.set_index('Date', inplace=True)
df=df.set_index('Date') # Set the DataFrame index using existing columns.
dfs.append( df ) # dfs[appl, pg, ...]
# Concatenate all dataframes into a single one
stock_df = pd.concat( dfs, axis=1 )
# Sort by ascending order of Company then Date
stock_df = stock_df.sort_index(axis=0)
stock_dfhttps://data.nasdaq.com/api/v3/datasets/WIKI/AAPL.json
"column_names":["Date","Open","High","Low","Close","Volume","Ex-Dividend","Split Ratio","Adj. Open","Adj. High","Adj. Low","Adj. Close","Adj. Volume"]
Let's normalize our dataset before using it for analysis:
https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.fillna.html
method{‘backfill’, ‘bfill’, ‘pad’, ‘ffill’, None}, default None
Method to use for filling holes in reindexed Series
- pad / ffill: propagate last valid observation forward to next valid
 ==>
==>
- backfill / bfill: use next valid observation to fill gap.
df_components[-20:]If you inspect every value in this data feed, you will notice NaN values, or missing data. Since we are using data that is error-prone, and for quick studies of PCA, we can temporarily fill in these unknown variables by propagating previous observed values. The fillna(method=' ffill' ) method helps to do this and stores the result in the filled_df_components variable.
filled_df_components = df_components.fillna(method='ffill' )
filled_df_components[-20:]An additional step in normalizing is to resample the time series at regular intervals and match it up exactly with our Dow time series dataset, which we will be downloading later. The daily_df_components variable stores the result from resampling the time series on a daily basis, and any missing values during resampling are propagated using the forward fill method. And finally, to account for incomplete starting data, we will simply perform a backfill of values with fillna(method=' bfill' )
daily_df_components = filled_df_components.resample('24h').ffill()
daily_df_components = daily_df_components.fillna(method='bfill' )
daily_df_components![]()
For the purpose of PCA demonstration, we have to make do with free, low-quality datasets. If you require high quality datasets, consider subscribing to a data publisher.
Downloading the DJIA dataset from quandl
start_date = '2000-01-01'
end_date = '2018-12-1'
stock_symbol='WIKI/NDAQ'
df_ndaq = quandl.get( stock_symbol,
start_date='2000-01-01',
end_date='2021-12-31',
)
df_ndaq.info()df_ndaqimport yfinance as yf
ndaq_df = yf.download( '^DJI', start=start_date, end=end_date)
ndaq_dfIt is clear that the data obtained by yfinance is more complete and accurate
import pandas as pd
df_ndaq=ndaq_df
# Prepare the dataframe
df_dji = pd.DataFrame( df_ndaq['Adj. Close'] )
df_dji.columns = ['DJIA']
df_dji.index = pd.to_datetime( df_dji.index )
# Trim the new dataframe and resample
djia_2017 = pd.DataFrame( df_dji.loc['2017-01-01':'2017-12-31'] )
djia_2017 = djia_2017.resample('24h').ffill()Here, we are taking the adjusted closing prices of Dow Jones for the year of 2017, resampled on a daily basis. The resulting DataFrame object is stored in djia_2017, which we can use for applying PCA.
Applying a kernel PCA
Principal Component Analysis - PCA
Similar to feature selection, we can use different feature extraction techniques to reduce the number of features in a dataset. The difference between feature selection and feature extraction is that while we maintain the original features when we used feature selection algorithms(select a subset of the original features), such as sequential backward selection, we use feature extraction to transform or project the data onto a new feature space(derive information from the feature set to construct a new feature subspace). In the context of dimensionality reduction, feature extraction can be understood as an approach to data compression with the goal of maintaining most of the relevant information. In practice, feature extraction is not only used to improve storage space or the computational efficiency of the learning algorithm, but can also improve the predictive performance by reducing the curse of dimensionality—especially if we are working with non-regularized models.
The Principal Component Analysis (PCA) aims to find combinations of features that describe the dataset in less information. It aims to discover principal components, which are features that do not correlate with each other and explain the information—specifically the variance—of the dataset. What this means is that we can often capture most of the information in a dataset in fewer features.
The main steps behind principal component analysis
In this section, we will discuss PCA (Principal Component Analysis), an unsupervised linear transformation technique that is widely used across different fields, most prominently for feature extraction and dimensionality reduction. Other popular applications of PCA include
exploratory data analyses and de-noising去噪 of signals in stock market trading, and the analysis of genome data and gene expression levels in the field of bioinformatics生物信息学.
PCA helps us to identify patterns in data based on the correlation between features. In a nutshell简言之, PCA aims to find the directions of maximum variance in highdimensional data and projects it onto a new subspace with equal or fewer dimensions than the original one(PCA identifies the hyperplane that lies closest to the data, and then it projects the data onto it.). The orthogonal axes (principal components) of the new subspace can be interpreted as the directions of maximum variance given the constraint that the new feature axes are orthogonal to each other, as illustrated in the following figure:![]() Here,
Here, ![]() and
and ![]() are the original feature axes, and PC1 and PC2 are the principal components.
are the original feature axes, and PC1 and PC2 are the principal components.
Preserving the Variance保留(最大)方差
![]() Figure 8-7. Selecting the subspace onto which to project https://blog.csdn.net/Linli522362242/article/details/105139547
Figure 8-7. Selecting the subspace onto which to project https://blog.csdn.net/Linli522362242/article/details/105139547
Before you can project the training set onto a lower-dimensional hyperplane, you first need to choose the right hyperplane. For example, a simple 2D dataset is represented on the left of Figure 8-7, along with three different axes (i.e., one-dimensional hyperplanes). On the right is the result of the projection of the dataset onto each of these axes. As you can see, the projection onto the solid line preserves the maximum variance, while the projection onto the dotted line preserves very little variance, and the projection onto the dashed line preserves an intermediate amount of variance. variance measures the spread of values along a feature axis(variance measures值沿特征轴的分布).
It seems reasonable to select the axis that preserves the maximum amount of variance(more values spread along the selected axis), as it will most likely lose less information than the other projections. Another way to justify this choice is that it is the axis that minimizes the mean squared distance between the original dataset and its projection onto that axis. This is the rather simple idea behind PCA.
################################
点积在数学中,又称数量积(dot product; scalar product)
a·b=(a^T)*b,这里的a^T指示矩阵a的转置。
![]()
正交矩阵
正交矩阵是在欧几里得空间里的叫法,在酉空间里叫酉矩阵,一个正交矩阵对应的变换叫正交变换,这个变换的特点是不改变向量的尺寸和向量间的夹角,那么它到底是个什么样的变换呢?看下面这张图![]()
假设二维空间中的一个向量OA,它在标准坐标系即向量e1、e2所在的坐标轴,坐标矩阵是![]() (用T表示转置), 现在把它用另一组向量e1'、e2'表示为
(用T表示转置), 现在把它用另一组向量e1'、e2'表示为![]() ,存在矩阵U使得
,存在矩阵U使得![]() ,则U即为正交矩阵。从图中可以看到,正交变换只是将变换向量用另一组正交基表示,在这个过程中并没有对向量OA做拉伸,也不改变向量OA的空间位置,加入两个向量同时做正交变换,那么变换前后这两个向量的夹角显然不会改变。上面的例子只是正交变换的一个方面,即旋转变换,可以把e1'、e2'坐标系看做是e1、e2坐标系经过旋转某个斯塔
,则U即为正交矩阵。从图中可以看到,正交变换只是将变换向量用另一组正交基表示,在这个过程中并没有对向量OA做拉伸,也不改变向量OA的空间位置,加入两个向量同时做正交变换,那么变换前后这两个向量的夹角显然不会改变。上面的例子只是正交变换的一个方面,即旋转变换,可以把e1'、e2'坐标系看做是e1、e2坐标系经过旋转某个斯塔![]() 角度得到,怎么样得到该旋转矩阵U呢?如下
角度得到,怎么样得到该旋转矩阵U呢?如下
向量OA:![]()
 OR ||
OR || ||= ||e1'|| * ||x|| * cosB 角度B是向量OA和单位向量
||= ||e1'|| * ||x|| * cosB 角度B是向量OA和单位向量 的夹角
的夹角 OR ||
OR || ||= ||e2'|| * ||x|| * cosC 角度C是向量OA和单位向量
||= ||e2'|| * ||x|| * cosC 角度C是向量OA和单位向量 的夹角
的夹角
![]() 和
和![]() 实际上是向量OA(or x)在e1'和e2'轴上的投影大小,所以直接做内积dot, then
实际上是向量OA(or x)在e1'和e2'轴上的投影大小,所以直接做内积dot, then ![]()
从图中可以看到 单位向量(模等于1的向量)![]() 和单位向量
和单位向量![]() 用向量e1、e2所在的坐标轴表示
用向量e1、e2所在的坐标轴表示![]() 所以
所以 ![]()
正交矩阵U行(列)向量之间都是单位正交向量。上面求得的是一个旋转矩阵,它对向量做旋转变换!向量OA空间位置空间位置不变是绝对的,但是坐标是相对的,假如你站在e1上看OA,随着e1旋转到e1',看OA的相对位置就会改变。
import matplotlib.pyplot as plt
import numpy as np
angle = np.pi/5
stretch = 5
m = 200
# create dataset
np.random.seed(3)
X = np.random.randn(m,2) /10 #randn: "n" is short for normal distribution
X = X.dot( np.array([ [stretch,0],
[0,1]
]
)
) #stretch # X = (x1*stretch, x2)
# Orthogonal matrix U
X = X.dot([ [np.cos(angle), np.sin(angle)],
[-np.sin(angle), np.cos(angle)]
]) # rotate
u1 = np.array([ np.cos(angle), np.sin(angle) ]) # c1
u2 = np.array([ np.cos(angle-2*np.pi/6), np.sin(angle-2*np.pi/6) ])
u3 = np.array([ np.cos(angle-np.pi/2), np.sin(angle-np.pi/2) ]) # c2
# X.dot(e1')
X_proj1 = X.dot( u1.reshape(-1,1) ) # u1.reshape(-1,1) hidden: u1.T Tranpose
X_proj2 = X.dot( u2.reshape(-1,1) )
# X.dot(e2')
X_proj3 = X.dot( u3.reshape(-1,1) )
plt.figure( figsize=(10,5) )
# shape : sequence of 2 ints ~ (3,2)
# Shape of grid in which to place axis.
# First entry is number of rows, second entry is number of columns.
# loc : sequence of 2 ints ~ (0,0)
# Location to place axis within grid.
# First entry is row number, second entry is column number.
plt.subplot2grid( (3,2), (0,0), rowspan=3 )
# c1
plt.plot( [-1.4, 1.4],
[ -1.4*u1[1]/u1[0], 1.4*u1[1]/u1[0] ],
"b-", linewidth=1
)
#
plt.plot( [-1.4, 1.4],
[ -1.4*u2[1]/u2[0], 1.4*u2[1]/u2[0] ],
"g--", linewidth=1
)
# c2
plt.plot( [-1.4, 1.4],
[ -1.4*u3[1]/u3[0], 1.4*u3[1]/u3[0] ],
"k:", linewidth=2
)
plt.plot( X[:,0], X[:,1], "bo", alpha=0.5 )
plt.axis([ -1.4,1.4, -1.4,1.4 ])
plt.arrow( 0,0, u1[0],u1[1],
head_width=0.1, linewidth=5, length_includes_head=True, head_length=0.1,
fc="k", ec="k")
plt.arrow( 0,0, u3[0],u3[1],
head_width=0.1, linewidth=5, length_includes_head=True, head_length=0.1,
fc="k", ec="k")
plt.text( u1[0]+0.1, u1[1]-0.05,
r"$\mathbf{c_1}$", fontsize=22 )
plt.text( u3[0]+0.1, u3[1],
r"$\mathbf{c_2}$", fontsize=22 )
plt.xlabel( "$x_1$", fontsize=18 )
plt.ylabel( "$x_2$", fontsize=18, rotation=0 )
plt.grid(True)
plt.subplot2grid( (3,2), (0,1) )
plt.plot( [-2,2], [0,0], "b-", linewidth=1 )
plt.plot( X_proj1[:,0], np.zeros(m), "bo", alpha=0.3 )
#plt.gca().get_yaxis().set_ticks([])
plt.gca().get_xaxis().set_ticklabels([])
plt.axis([-2,2, -1,1])
plt.grid(True)
plt.subplot2grid( (3,2), (1,1) )
plt.plot( [-2,2], [0,0], "g--", linewidth=1 )
plt.plot( X_proj2[:,0], np.zeros(m), "bo", alpha=0.3 )
plt.gca().get_yaxis().set_ticks([])
plt.gca().get_xaxis().set_ticklabels([])
plt.axis([-2,2,-1,1])
plt.grid(True)
plt.subplot2grid( (3,2), (2,1))
plt.plot( [-2,2], [0,0], "k:", linewidth=2 )
plt.plot( X_proj3[:,0], np.zeros(m), "bo", alpha=0.3 )
plt.gca().get_yaxis().set_ticks([])
#plt.gca().get_xaxis().set_ticklabels([])
plt.axis([-2,2,-1,1])
plt.xlabel("$z_1$", fontsize=18)
plt.grid(True)
plt.show() ![]()
NOTE
The direction of the principal components is not stable: if you perturb打乱 the training set slightly and run PCA again, some of the new PCs(Principal Components) may point in the opposite direction of the original PCs. However, they will generally still lie on the same axes. In some cases, a pair of PCs may even rotate or swap, but the plane they define will generally remain the same.
################################
If we use PCA for dimensionality reduction, we construct a ![]() –dimensional transformation matrix W that allows us to map a sample vector x onto a new k–dimensional feature subspace that has fewer dimensions than the original d–dimensional feature space(k
–dimensional transformation matrix W that allows us to map a sample vector x onto a new k–dimensional feature subspace that has fewer dimensions than the original d–dimensional feature space(k![]()
![]()
![]()
As a result of transforming the original d-dimensional data onto this new k-dimensional subspace (typically k << d), the first principal component will have the largest possible variance, and all consequent principal components will have the largest variance given the constraint that these components are uncorrelated (orthogonal) to the other principal components—even if the input features are correlated, the resulting principal components will be mutually orthogonal (uncorrelated). Note that the PCA directions are highly sensitive to data scaling, and we need to standardize the features prior to PCA if the features were measured on different scales and we want to assign equal importance to all features.
Before looking at the PCA algorithm for dimensionality reduction in more detail, let's summarize the approach in a few simple steps:
- Standardize the d-dimensional dataset
 .
.

 ==>
==>
- Construct the covariance matrix A (dxd).

(the correlation matrix is identical to a covariance matrix
is identical to a covariance matrix computed from standardized features
computed from standardized features ==>
==> ==>
==> <==
<==
Finally, we can simplify this equation as follows: ( correlation coefficient formula)
( correlation coefficient formula)
https://blog.csdn.net/Linli522362242/article/details/120398175
)
https://blog.csdn.net/Linli522362242/article/details/111307026
Covariance is a measure of how much two variables change together and it is a measure of the strength of the correlation between two sets of variables. If the covariance of two variables is zero, we can conclude that there will not be any correlation between two sets of the variables. The formula for covariance is as follows: vs population covariances
vs population covariances
A sample covariance calculation is shown for X and Y variables in the following formulas. However, it is a 2 x 2 matrix of an entire covariance matrix (also, it is a square matrix).
- Decompose分解 the covariance matrix into its eigenvectors(non-zero vectors) and eigenvalues.
 ==>
==>
################### ==>
==> ==>
==>
 ==>
==>
==> ==>
==>
 ==>divided by
==>divided by ==>unit eigenvectors
==>unit eigenvectors
###################import numpy as np # Covariance matrix A eigen_vals, eigen_vecs = np.linalg.eig( np.array([ [0.91335, 0.75969], [0.75969, 0.69702] ]) ) print( '\nEigen Values\n', eigen_vals ) print( '\nEigen Vectors\n', eigen_vecs ) OR
OR
- Sort the eigenvalues by decreasing order to rank the corresponding eigenvectors.
- Select k eigenvectors which correspond to the k largest eigenvalues, where k is the dimensionality of the new feature subspace (
 ).
). - Construct a projection matrix W from the "top" k eigenvectors.

- Transform the d-dimensional input dataset X using the projection matrix W to obtain the new k-dimensional feature subspace.
 From the preceding result, we can see the 1D projection of the first principal component from the original 2D data. Also, the eigenvalue of 1.57253666 explains the fact that the principal component explains variance of 57 percent more than the original variables.
From the preceding result, we can see the 1D projection of the first principal component from the original 2D data. Also, the eigenvalue of 1.57253666 explains the fact that the principal component explains variance of 57 percent more than the original variables.
In the case of multi-dimensional data, the thumb rule is to select the eigenvalues or principal components(eigenvectors) with a value greater than what should be considered for projection.
Eigenvectors and eigenvalues
Eigenvectors and eigenvalues have significant importance in the field of linear algebra, physics, mechanics, and so on. Refreshing, basics on eigenvectors and eigenvalues is necessary when studying PCAs. Eigenvectors![]() are the axes (directions) along which a linear transformation acts simply by stretching/compressing and/or flipping; whereas, eigenvalues λ give you the factors by which the compression occurs. In another way, an eigenvector of a linear transformation is a nonzero vector whose direction does not change when that linear transformation is applied to it.
are the axes (directions) along which a linear transformation acts simply by stretching/compressing and/or flipping; whereas, eigenvalues λ give you the factors by which the compression occurs. In another way, an eigenvector of a linear transformation is a nonzero vector whose direction does not change when that linear transformation is applied to it.
More formally, A is a linear transformation from a vector space and ![]() is a nonzero vector, then eigen vector of A if
is a nonzero vector, then eigen vector of A if ![]() is a scalar multiple of
is a scalar multiple of ![]() . The condition can be written as the following equation:
. The condition can be written as the following equation:![]()
In the preceding equation, ![]() is an eigenvector, A is a square matrix, and λ is a scalar called an eigenvalue. The direction of an eigenvector remains the same after it has been transformed by A; only its magnitude has changed, as indicated by the eigenvalue λ, That is,
is an eigenvector, A is a square matrix, and λ is a scalar called an eigenvalue. The direction of an eigenvector remains the same after it has been transformed by A; only its magnitude has changed, as indicated by the eigenvalue λ, That is,![]() multiplying a matrix by one of its eigenvectors is equal to scaling the eigenvector, which is a compact representation of the original matrix. The following graph describes eigenvectors and eigenvalues in a graphical representation in a 2D space
multiplying a matrix by one of its eigenvectors is equal to scaling the eigenvector, which is a compact representation of the original matrix. The following graph describes eigenvectors and eigenvalues in a graphical representation in a 2D space
![]()
The following example describes how to calculate eigenvectors and eigenvalues from the square matrix(a linear transformation) and its understanding. Note that eigenvectors and eigenvalues can be calculated only for square matrices (those with the same dimensions of rows and columns).![]()
Recall the equation![]() that the product of A and any eigenvector of A must be equal to the eigenvector multiplied by the magnitude of eigenvalue:
that the product of A and any eigenvector of A must be equal to the eigenvector multiplied by the magnitude of eigenvalue:![]() ==>
==>![]()
![]() (通过行列式必须为0 得到特征值)
(通过行列式必须为0 得到特征值)
A characteristic equation特征方程 states that the determinant行列式 of the matrix, that is the difference between the data matrix and the product of the identity matrix单位矩阵 and an eigenvalue is 0.
Both eigenvalues λ for the preceding matrix are equal to -2. We can use eigenvalues λ to substitute for eigenvectors![]() in an equation:
in an equation:
![]()
![]()
Substituting the value of eigenvalue in the preceding equation, we will obtain the following formula:![]()
The preceding equation can be rewritten as a system of equations, as follows: ![]()
This equation indicates it can have multiple solutions of eigenvectors we can substitute with any values which hold the preceding equation for verification of equation. Here, we have used the vector [1 1] for verification, which seems to be proved.![]()
![]() and eigenvalues λ = -2
and eigenvalues λ = -2
PCA needs unit eigenvectors![]() to be used in calculations, hence we need to divide the same with the norm or we need to normalize the eigenvector. The 2-norm equation is shown as follows:
to be used in calculations, hence we need to divide the same with the norm or we need to normalize the eigenvector. The 2-norm equation is shown as follows:![]() OR
OR ![]()
The norm of the output vector is calculated as follows:![]()
The unit eigenvector is shown as follows:![]()
#####################
Specifically, every real symmetric matrix can be decomposed into an expression using only real-valued eigenvectors and eigenvalues: ![]() (2.41)
(2.41)
where ![]() is an orthogonal matrix composed of eigenvectors of A(A has n linearly independent eigenvectors), and
is an orthogonal matrix composed of eigenvectors of A(A has n linearly independent eigenvectors), and ![]() is a diagonal matrix(n个特征值为主对角线的n×n维矩阵). The eigenvalue
is a diagonal matrix(n个特征值为主对角线的n×n维矩阵). The eigenvalue ![]() is associated with the eigenvector in column
is associated with the eigenvector in column ![]() of Q, denoted as
of Q, denoted as ![]() . Because Q is an orthogonal matrix, we can think of A as scaling space by
. Because Q is an orthogonal matrix, we can think of A as scaling space by ![]() in direction
in direction ![]() . See figure 2.3 for an example.
. See figure 2.3 for an example. Figure 2.3: An example of the effect of eigenvectors and eigenvalues. Here, we have a matrix A with two orthonormal eigenvectors,
Figure 2.3: An example of the effect of eigenvectors and eigenvalues. Here, we have a matrix A with two orthonormal eigenvectors,
 with eigenvalue
with eigenvalue  and
and with eigenvalue
with eigenvalue  .
.- (Left)We plot the set of all unit vectors u ∈
 as a unit circle.
as a unit circle. - (Right)We plot the set of all points Au. By observing the way that A distorts/ dɪˈstɔːrt /拉伸,扭曲,变形 the unit circle, we can see that it scales space in direction
 by
by  .
.
While any real symmetric matrix A is guaranteed to have an eigendecomposition, the eigendecomposition may not be unique. If any two or more eigenvectors share the same eigenvalue, then any set of orthogonal vectors lying in their span are also eigenvectors with that eigenvalue(如果两个或多个特征向量拥有相同的特征值,那么在由这些特征向量生成的子空间中,任意一组正交向量都是该特征值对应的特征向量), and we could equivalently choose a Q using those eigenvectors instead. By convention, we usually sort the entries of ![]() in descending order. Under this convention, the eigendecomposition is unique only if all of the eigenvalues are unique.
in descending order. Under this convention, the eigendecomposition is unique only if all of the eigenvalues are unique.
The eigendecomposition of a matrix tells us many useful facts about the matrix. The matrix is singular if and only if any of the eigenvalues are zero. The eigendecomposition of a real symmetric matrix can also be used to optimize quadratic expressions of the form ![]() subject to
subject to  . Whenever
. Whenever ![]() is equal to an eigenvector of
is equal to an eigenvector of ![]() ,
, ![]() takes on the value of the corresponding eigenvalue. The maximum value of
takes on the value of the corresponding eigenvalue. The maximum value of ![]() within the constraint region is the maximum eigenvalue and its minimum value within the constraint region is the minimum eigenvalue.
within the constraint region is the maximum eigenvalue and its minimum value within the constraint region is the minimum eigenvalue.
- A matrix whose eigenvalues are all positive is called positive definite正定.
- A matrix whose eigenvalues are all positive or zero-valued is called positive semidefinite半正定.
- Likewise, if all eigenvalues are negative, the matrix is negative definite负定, and
- if all eigenvalues are negative or zero-valued, it is negative semidefinite半负定.
- Positive semidefinite matrices are interesting because they guarantee that ∀x,
 . Positive definite matrices additionally guarantee that
. Positive definite matrices additionally guarantee that  ⇒ x = 0.
⇒ x = 0.
1. 回顾特征值和特征向量
我们首先回顾下特征值和特征向量的定义如下:Ax=λx (hidden: ==>左乘x' ==> A = x'λx)

其中A是一个n×n的实对称矩阵(如果有n阶矩阵A,其矩阵的元素都为实数,且矩阵A的转置等于其本身( ),(i,j为元素的脚标),则称A为实对称矩阵。)
),(i,j为元素的脚标),则称A为实对称矩阵。)- 实对称矩阵A的 不同特征值 对应的 特征向量 是正交的。
- 实对称矩阵A的特征值都是实数。
- n阶实对称矩阵A必可相似对角化,且相似对角矩阵
 上的元素即为矩阵本身特征值。
上的元素即为矩阵本身特征值。 - 若A具有 k重特征值λ 必有k个线性无关的特征向量,或者说秩r
 必为n-k,其中
必为n-k,其中 为单位矩阵。
为单位矩阵。 - 实对称矩阵A一定可正交相似对角化。


- x是一个n维向量,则我们说λ是矩阵A的一个特征值,而x 是 矩阵A 的 特征值λ 所对应的特征向量.
求出特征值和特征向量有什么好处呢? 就是我们可以将矩阵A特征分解。如果我们求出了矩阵A的n个特征值 λ1 ≤ λ2 ≤...≤ λn,以及这n个特征值所对应的特征向量{w1,w2,...wn} ,注意 wi是n维的 ,如果这n个特征向量线性无关(线性无关一般指线性独立。 线性独立一般是指向量的线性独立,指一组向量中任意一个向量都不能由其它几个向量线性表示),那么矩阵A就可以用下式的特征分解表示:![]() or
or ![]()
其中W是这n个特征向量{w1,w2,...wn}所组成的n×n维矩阵,而Σ为这n个特征值为主对角线的n×n维矩阵。
一般我们会把W的这n个特征向量标准化,即满足![]() , 或者说
, 或者说![]() ,此时W的n个特征向量为标准正交基,满足
,此时W的n个特征向量为标准正交基,满足![]() ,即
,即![]() , 也就是说W为酉矩阵。
, 也就是说W为酉矩阵。
这样我们的特征分解表达式可以写成![]()
注意到要进行特征分解,矩阵A必须为方阵。那么如果A不是方阵,即行和列不相同时,我们还可以对矩阵进行分解吗?答案是可以,此时我们的SVD登场了。
2. SVD的定义 奇异值分解(singular value decomposition)
we saw how to decompose a matrix into eigenvectors and eigenvalues. The singular value decomposition (SVD) provides another way to factorize a matrix, into singular vectors and singular values. The SVD allows us to discover some of the same kind of information as the eigendecomposition. However, the SVD is more generally applicable. Every real matrix has a singular value decomposition, but the same is not true of the eigenvalue decomposition. For example, if a matrix is not square, the eigendecomposition is not defined, and we
must use a singular value decomposition instead.
Recall that the eigendecomposition involves analyzing a matrix A to discover a matrix V of eigenvectors and a vector of eigenvalues λ such that we can rewrite A as![]()
The singular value decomposition is similar, except this time we will write A as a product of three matrices: ![]() OR
OR ![]()
- A is an m×n matrix.
- Then U is defined to be an m×m matrix,
The columns of U are known as the left-singular vectors - D OR Σ to be an m× n matrix,
The matrix D OR Σ is defined to be a diagonal matrix. Note that D OR Σ is not necessarily square.
The elements along the diagonal of D OR Σ are known as the singular values of the matrix A.
除了主对角线上的元素以外全为0,主对角线上的每个元素都称为奇异值, - and V to be an n × n matrix.
The columns of V are known as as the right-singular vectors. - The matrices U and V are both defined to be orthogonal matrices.
U和V都是酉矩阵(正交矩阵是在欧几里得空间里的叫法,在酉空间里叫酉矩阵),即满足 。下图可以很形象的看出上面SVD的定义:
。下图可以很形象的看出上面SVD的定义:
那么我们如何求出SVD分解后的U,Σ,V这三个矩阵呢?
- 如果我们将A的转置和A 做矩阵乘法,那么会得到n×n的一个方阵
 (nxm * mxn=nxn维矩阵)。既然是方阵,那么我们就可以进行特征分解,得到的特征值和特征向量满足下式: (similar to Ax=λx)
(nxm * mxn=nxn维矩阵)。既然是方阵,那么我们就可以进行特征分解,得到的特征值和特征向量满足下式: (similar to Ax=λx)
这样我们就可以得到矩阵的n个特征值和对应的n个特征向量了。将的所有特征向量组成一个n×n的矩阵V,就是我们SVD公式里面的V矩阵了。一般我们将V中的每个特征向量叫做A的右奇异向量。 (similar to Ax=λx)
(similar to Ax=λx)
这样我们就可以得到矩阵 的n个特征值
的n个特征值 和对应的n个特征向量
和对应的n个特征向量 了。将
了。将 的所有特征向量
的所有特征向量 组成一个n×n的矩阵V,就是我们SVD公式里面的V矩阵了。一般我们将V中的每个特征向量叫做A的右奇异向量(right sigular vector)。
组成一个n×n的矩阵V,就是我们SVD公式里面的V矩阵了。一般我们将V中的每个特征向量叫做A的右奇异向量(right sigular vector)。 - 如果我们将A和A的转置 做矩阵乘法,那么会得到m×m的一个方阵
 (mxn * nxm = mxm维矩阵)。既然
(mxn * nxm = mxm维矩阵)。既然 是方阵,那么我们就可以进行特征分解,得到的特征值和特征向量满足下式:
是方阵,那么我们就可以进行特征分解,得到的特征值和特征向量满足下式:
这样我们就可以得到矩阵的m个特征值和对应的m个特征向量了。将的所有特征向量组成一个m×m的矩阵U,就是我们SVD公式里面的U矩阵了。一般我们将U中的每个特征向量叫做A的左奇异向量(left singular vector)。 - U和V我们都求出来了,现在就剩下奇异值矩阵Σ没有求出了。由于Σ除了对角线上是奇异值其他位置都是0,那我们只需要求出每个奇异值σ就可以了。

我们注意到:
这样我们可以求出我们的每个奇异值,进而求出奇异值矩阵Σ。 - 上面还有一个问题没有讲,
 就是我们说
就是我们说 的特征向量
的特征向量 组成的就是我们SVD中的V矩阵,
组成的就是我们SVD中的V矩阵, 而
而 的特征向量
的特征向量 组成的就是我们SVD中的U矩阵,
组成的就是我们SVD中的U矩阵,
这有什么根据吗?这个其实很容易证明,我们以V矩阵的证明为例
上式证明使用了: 。
。
可以看出 的特征向量组成的的确就是我们SVD中的V矩阵(hidden: Ax=λx ==> x 是 矩阵A 的 特征值λ 所对应的特征向量 ==>左乘x' ==> A = x'λx )。类似的方法可以得到
的特征向量组成的的确就是我们SVD中的V矩阵(hidden: Ax=λx ==> x 是 矩阵A 的 特征值λ 所对应的特征向量 ==>左乘x' ==> A = x'λx )。类似的方法可以得到 的特征向量组成的就是我们SVD中的U矩阵
的特征向量组成的就是我们SVD中的U矩阵
进一步我们还可以看出我们的特征值矩阵
 等于奇异值矩阵Σ的平方,也就是说特征值和奇异值满足如下关系:
等于奇异值矩阵Σ的平方,也就是说特征值和奇异值满足如下关系: ==>
==> =
= ( and
( and  ) ==>
) ==>
这样也就是说,我们可以不用 (<==
(<== )来计算奇异值,也可以通过求出
)来计算奇异值,也可以通过求出 的特征值取平方根来求奇异值。
的特征值取平方根来求奇异值。
3. SVD计算举例
![]()
进而求出![]() 的特征值和特征向量
的特征值和特征向量![]() :
:![]()
![]() ==>
==>![]() 特征向量标准化
特征向量标准化 ![]() =1 OR
=1 OR ![]() =1
=1![]() (特征值矩阵
(特征值矩阵![]() 等于奇异值矩阵Σ的平方)
等于奇异值矩阵Σ的平方)
![]() 特征值取平方根来求奇异值==>
特征值取平方根来求奇异值==>![]() ,
, ![]()
![]()
接着求![]() 的特征值和特征向量
的特征值和特征向量![]() :过程与求出
:过程与求出![]() 的特征值和特征向量
的特征值和特征向量![]() 类似:
类似:![]()
![]()
![]()
![]()
![]()
![]()
So how can you find the principal components of a training set? Luckily, there is a standard matrix factorization因数分解 technique called Singular Value Decomposition (SVD)奇异值分解 that can decompose the training set matrix X into the dot product of three matrices![]() where contains all the principal components that we are looking for, as shown in Equation 8-1.
where contains all the principal components that we are looking for, as shown in Equation 8-1.
Equation 8-1. Principal components matrix![]()
https://blog.csdn.net/Linli522362242/article/details/120398175######################################
Kernel PCA) Using kernel principal component analysis for nonlinear mappings
Many machine learning algorithms make assumptions about the linear separability of the input data. You learned that the perceptron even requires perfectly linearly separable training data to converge. Other algorithms that we have covered so far assume that the lack of perfect linear separability is due to noise: Adaline, logistic regression, and the (standard) support vector machine (SVM) to just name a few只举几个.
However, if we are dealing with nonlinear problems, which we may encounter rather frequently in real-world applications, linear transformation techniques for dimensionality reduction, such as PCA and LDA, may not be the best choice. In this section, we will take a look at a kernelized version of PCA, or KPCA, which relates to the concepts of kernel SVM that we remember from Cp3, A Tour of Machine Learning Classifiers Using scikit-learn (https://blog.csdn.net/Linli522362242/article/details/96480059). Using kernel PCA, we will learn how to transform data that is not linearly separable onto a new, lower-dimensional subspace that is suitable for linear classifier
![]()
Kernel functions and the kernel trick
As we remember from our discussion about kernel SVMs in Cp3https://blog.csdn.net/Linli522362242/article/details/96480059, A Tour of Machine Learning Classifiers Using Scikit-learn, we can tackle nonlinear problems by projecting them onto a new feature space of higher dimensionality where the classes become linearly separable. To transform the samples ![]() onto this higher k-dimensional subspace, we defined a nonlinear mapping function
onto this higher k-dimensional subspace, we defined a nonlinear mapping function![]() :
:![]()
We can think of ![]() as a function that creates nonlinear combinations of the original features to map the original d-dimensional dataset onto a larger, k-dimensional feature space. For example, if we had feature vector
as a function that creates nonlinear combinations of the original features to map the original d-dimensional dataset onto a larger, k-dimensional feature space. For example, if we had feature vector![]() (
( ![]() is a column vector consisting of d features) with two dimensions (d = 2) , a potential mapping onto a 3D space could be as follows:
is a column vector consisting of d features) with two dimensions (d = 2) , a potential mapping onto a 3D space could be as follows:![]()
In other words, via kernel PCA we perform a nonlinear mapping that transforms the data onto ==> a higher-dimensional space and use standard PCA in this higher-dimensional space to project the data back onto==> a lower-dimensional space where the samples can be separated by a linear classifier (under the condition that the samples can be separated by density in the input space). However, one downside of this approach is that it is computationally very expensive, and this is where we use the kernel trick.
Using the kernel trick, we can compute the similarity between two high-dimension feature vectors in the original feature space.
Before we proceed with more details about the kernel trick to tackle this computationally expensive problem, let us think back to the standard PCA approach that we implemented at the beginning of this chapter. We computed the covariance between two features k and j as follows:
Example:
a covariance matrix of three features can then be written as (note that ![]() stands for the Greek uppercase letter sigma, which is not to be confused with the sum symbol):
stands for the Greek uppercase letter sigma, which is not to be confused with the sum symbol):![]()
Since the standardizing of features centers them at mean zero, for instance,![]() and
and ![]() , we can simplify this equation
, we can simplify this equation  as follows:
as follows:
Note that the preceding equation refers to the covariance between two features; now, let's write the general equation to calculate the covariance matrix ![]() :
:
and
( ![]() is a column vector consisting of d features (d-dimenional vector),
is a column vector consisting of d features (d-dimenional vector), ![]() is the sample instance index and
is the sample instance index and ![]() ),
),
Bernhard Scholkopf generalized this approach (B. Scholkopf, A. Smola, and K.-R. Muller. Kernel Principal Component Analysis. pages 583–588, 1997) so that we can replace the dot products between samples in the original feature space by the nonlinear feature combinations via ![]() :
:![]()
To obtain the eigenvectors—the principal components—from this covariance matrix, we have to solve the following equation:
######################
https://blog.csdn.net/Linli522362242/article/details/105139547
12.1.1 Maximum variance formulation
Consider a data set of observations![]() where n = 1, . . . , N, and
where n = 1, . . . , N, and![]() is a Euclidean variable with dimensionality D (D features). Our goal is to project the data onto a space having dimensionality M
is a Euclidean variable with dimensionality D (D features). Our goal is to project the data onto a space having dimensionality M
To begin with, consider the projection onto a one-dimensional space (M = 1). We can define the direction of this space using a D-dimensional vector![]() (
(![]() ), which for convenience (and without loss of generality) we shall choose to be a unit vector so that
), which for convenience (and without loss of generality) we shall choose to be a unit vector so that![]() = 1 (note that we are only interested in the direction defined by
= 1 (note that we are only interested in the direction defined by![]() , not in the magnitude of itself).
, not in the magnitude of itself).
- Each data point
 is then projected onto a scalar value
is then projected onto a scalar value  .
. - The mean of the projected data is
 (
( ) where
) where  is the sample set mean given by
is the sample set mean given by (12.1)
(12.1)
Note (=
(= ) has D dimensions (D features), and the inner
) has D dimensions (D features), and the inner  is the mean for each feature
is the mean for each feature- and the variance of the projected data is given by
 (12.2
(12.2
hide:


- where S is the data covariance 协方差 matrix defined by
 (12.3)
(12.3)
We now maximize the projected variance![]() with respect to
with respect to![]() . Clearly, this has to be a constrained maximization to prevent
. Clearly, this has to be a constrained maximization to prevent ![]() . The appropriate constraint comes from the normalization condition归一化条件
. The appropriate constraint comes from the normalization condition归一化条件 ![]() = 1. To enforce this constraint, we introduce a Lagrange multiplier that we shall denote by
= 1. To enforce this constraint, we introduce a Lagrange multiplier that we shall denote by ![]() , and then make an unconstrained maximization of
, and then make an unconstrained maximization of ![]() (12.4)
(12.4)
By setting the derivative with respect to![]() equal to zero, we see that this quantity will have a stationary point驻点 when
equal to zero, we see that this quantity will have a stationary point驻点 when![]() (12.5
(12.5![]() ) which says that
) which says that![]() must be an eigenvector特征向量 of S. If we left-multiply by
must be an eigenvector特征向量 of S. If we left-multiply by![]() and make use of
and make use of![]() = 1, we see that the variance is given by
= 1, we see that the variance is given by ![]() (12.6
(12.6![]() )
)
and so the variance will be a maximum when we set![]() equal to the eigenvector having the largest eigenvalue
equal to the eigenvector having the largest eigenvalue![]() . This eigenvector is known as the first principal component.
. This eigenvector is known as the first principal component.
We can define additional principal components in an incremental fashion方式 by choosing each new direction to be that which maximizes the projected variance amongst all possible directions orthogonal to those already considered. If we consider the general case of an M-dimensional projection space, the optimal linear projection for which the variance of the projected data is maximized is now defined by the M eigenvectors![]() of the data covariance matrix S corresponding to the M largest eigenvalues
of the data covariance matrix S corresponding to the M largest eigenvalues![]() . This is easily shown using proof by induction.
. This is easily shown using proof by induction.
######################
Since the standardizing of features centers them at mean zero, for instance,![]() and
and ![]() , we can simplify this equation
, we can simplify this equation  as follows:
as follows:
 and
and==>
![]() ==>
==>![]()


(Note :![]()

标量(scalar),亦称“无向量”。有些物理量,只具有数值大小,而没有方向,部分有正负之分。物理学中,标量(或作纯量)指在坐标变换下保持不变的物理量。用通俗的说法,标量是只有大小,没有方向的量
the eigenvectors can be written as a linear combination for features(1<= j<= k features ) : <==
<==![]() is the feature value and i=1
is the feature value and i=1![]()
)
Here, ![]() and
and![]() are the eigenvalues and eigenvectors of the covariance matrix
are the eigenvalues and eigenvectors of the covariance matrix ![]() , and
, and![]() can be obtained by extracting the eigenvectors of the kernel (similarity) matrix K as we will see in the following paragraphs.
can be obtained by extracting the eigenvectors of the kernel (similarity) matrix K as we will see in the following paragraphs.
The derivation of the kernel matrix is as follows:
- First, let's write the covariance matrix as in matrix notation, where
 is an n×k -dimensional matrix:
is an n×k -dimensional matrix:
note: is k-dimenional vector (k features = previous d features)
is k-dimenional vector (k features = previous d features) - Now, we can write the eigenvector equation as follows:

Since , we get:
, we get:

Multiplying it by on both sides yields the following result: ###
on both sides yields the following result: ### 

Here, K is the similarity (kernel) matrix:
https://www.cs.mcgill.ca/~dprecup/courses/ML/Lectures/ml-lecture13.pdf
-
As we recall from the SVM section in https://blog.csdn.net/Linli522362242/article/details/107755405, A Tour of Machine Learning Classifiers Using Scikit-learn, we use the kernel trick to avoid calculating the pairwise dot products of the samples
 under
under explicitly by using a kernel function K so that we don't need to calculate the eigenvectors explicitly:
explicitly by using a kernel function K so that we don't need to calculate the eigenvectors explicitly:
In other words, what we obtain after kernel PCA are the samples already projected onto the respective components在 kernel PCA 之后获得的是已经投影到各个components的样本 rather than constructing a transformation matrix as in the standard PCA approach 而不是像standard PCA方法中那样构建变换矩阵
(
for example
Construct a projection matrix W from the "top" k eigenvectors.
Transform the d-dimensional input dataset X using the projection matrix W to obtain the new k-dimensional feature subspace.
). Basically, the kernel function (or simply kernel) can be understood as a function that calculates a dot product between two vectors—a measure of similarity相似度.
The most commonly used kernels are as follows:
- The polynomial kernel:

Here, is the threshold and p is the power that has to be specified by the user.
is the threshold and p is the power that has to be specified by the user. - The hyperbolic tangent双曲线的正切 (sigmoid) kernel:

- The Radial Basis Function (RBF) or Gaussian kernel that we will use in the following examples in the next subsection:


 :
:
For example, if our dataset contains 100 training samples, the symmetric kernel matrix of the pair-wise similarities would be 100×100 dimensional.
To summarize what we have discussed so far, we can define the following three steps to implement an RBF kernel PCA:
- We compute the kernel (similarity) matrix k , where we need to calculate the following:

We do this for each pair of samples: note: is k-dimenional vector(k features)
is k-dimenional vector(k features)
For example, if our dataset contains n=100 training samples, the symmetric kernel matrix of the pair-wise similarities would be 100×100 dimensional. - We center the kernel matrix k using the following equation:

Here, is an n× n - dimensional matrix (the same dimensions as the kernel matrix) where all values are equal to
is an n× n - dimensional matrix (the same dimensions as the kernel matrix) where all values are equal to  .
.
#########################

 eigenvalue
eigenvalue
We have a normalization condition( ) for the vectors:
) for the vectors:
 标量
标量 是只有大小,没有方向的量
是只有大小,没有方向的量
By multiplying by
by  and using the normalization condition we get:
and using the normalization condition we get:


For a new point x, its projection onto the principal components is:
In general, may not be zero mean
may not be zero mean
Centered features:
The corresponding kernel is:
is: 

In a matrix form
where is a matrix with all elements
is a matrix with all elements .
.
OR
Here, is an n× n - dimensional matrix (the same dimensions as the kernel matrix) where all values are equal to
is an n× n - dimensional matrix (the same dimensions as the kernel matrix) where all values are equal to  .
.
######################### - We collect the top k eigenvectors of the centered kernel matrix based on their corresponding eigenvalues, which are ranked by decreasing magnitude. In contrast to standard PCA, the eigenvectors are not the principal component axes but the samples projected onto those axes.
At this point, you may be wondering why we need to center the kernel matrix in the 2nd step. We previously assumed that we are working with standardized data, where all features have mean zero(![]() <==
<==![]() ,
,![]() and
and![]() ) when we formulated the covariance matrix(
) when we formulated the covariance matrix(![]() ) and replaced the dot products(
) and replaced the dot products(![]() ) by the nonlinear feature combinations via
) by the nonlinear feature combinations via![]() .Thus, the centering of the kernel matrix in the second step becomes necessary, since we do not compute the new feature space explicitly and we cannot guarantee that the new feature space is also centered at zero.
.Thus, the centering of the kernel matrix in the second step becomes necessary, since we do not compute the new feature space explicitly and we cannot guarantee that the new feature space is also centered at zero.
https://blog.csdn.net/Linli522362242/article/details/105196037
Finding eigenvectors and eigenvalues
We can perform a kernel PCA using the KernelPCA class of the sklearn.decomposition module in Python. The default kernel method is linear. The dataset that's used in PCA is required to be normalized, which we can perform with z-scoring. The following code do this:
from sklearn.decomposition import KernelPCA
# https://blog.csdn.net/Linli522362242/article/details/108230328
# min-max scaling : (x-x.mean)/(x.max-x.min)
# standardization : ( x-x.mean() ) / x.std()
# https://blog.csdn.net/Linli522362242/article/details/121721868
fn_z_score = lambda x: ( x-x.mean() ) / x.std()
df_z_components = daily_df_components.apply( fn_z_score )
kpca = KernelPCA()
df_z_components_transformed = kpca.fit_transform( df_z_components)The fn_z_score variable is an inline function to perform z-scoring on a pandas DataFrame, which is applied with the apply() method. These normalized datasets can be fitted into a kernel PCA with the fit() method. The fitted results of the daily Dow component prices are stored in the df_z_components_transformed vallue, which is of the same KernelPCA object.
kpca.get_params()kernel PCA with linear kernel is exactly equivalent to the standard PCA.  https://axon.cs.byu.edu/~martinez/classes/778/Papers/KernelPCA.pdf
https://axon.cs.byu.edu/~martinez/classes/778/Papers/KernelPCA.pdf
parameters:
n_components int, default=None
Number of components. If None, all non-zero components are kept.
daily_df_components# standardization : ( x-x.mean() ) / x.std()
df_z_componentspd.DataFrame( df_z_components_transformed )
Two main outputs of PCA are eigenvectors and eigenvalues. Eigenvectors are vectors containing the direction of the principal component line, which doesn't change when a linear transformation is applied. Eigenvalues are scalar values indicating the amount of variance of the data in a direction with respect to a particular eigenvector( the largest variance can capture most of the information in a dataset). In fact, the eigenvector with the highest eigenvalue forms the principal component.
kpca.eigenvectors_![]()
kpca.eigenvectors_.shape![]()
(Standard PCA = linear kernel PCA) the eigenvectors are the principal component axes
kpca.eigenvalues_%matplotlib inline
import matplotlib.pyplot as plt
plt.figure( figsize=(12,8) )
plt.plot( kpca.eigenvalues_ )
plt.ylabel( 'eigenvalues' )
plt.show()
We can see that the first few eigenvalues explain much of the variances in the data, and become more negligent further down the components. Taking the first five eigenvalues, let's see how much explanation each of these eigenvalues gives us by obtaining their weighted average values:
The variance explained ratio(方差解释比率或者方差贡献率) of an eigenvalue is simply the fraction of an eigenvalue and the total sum of the eigenvalues:![]() https://blog.csdn.net/Linli522362242/article/details/120559394
https://blog.csdn.net/Linli522362242/article/details/120559394
# The amount of variance explained by each of the selected components. The variance estimation uses n_samples-1 degrees of freedom.
fn_weighted_avg = lambda x: x/x.sum()
weighted_values = fn_weighted_avg( kpca.eigenvalues_ ) [:5]
print( weighted_values ) ![]()
We can see that the 1st component explains 65% of the variance of the data, the 2nd component explains 14%, and so on. Taking the sum of these values, we get the following:
weighted_values.sum()![]() The first five eigenvalues would explain 92% of the variance in the dataset.
The first five eigenvalues would explain 92% of the variance in the dataset.
Reconstructing the Dow index with PCA
By default, the KernelPCA is instantiated with the n_components=None parameter, which constructs a kernel PCA with non-zero components. We can also create a PCA index with five components:
import numpy as np
# df_z_components = daily_df_components.apply( fn_z_score )
kernel_pca = KernelPCA( n_components=5 ).fit( df_z_components )
transformed_pca_5 = kernel_pca.transform( df_z_components )
pd.DataFrame( transformed_pca_5 )![]() 361x30 ==>map==> 361x5
361x30 ==>map==> 361x5
With the fit() method, we fitted the normalized dataset using the linear kernel PCA function with five components. The transform() method transforms the original dataset with the kernel PCA.
kernel_pca.eigenvalues_ ![]()
# fn_weighted_avg = lambda x: x/x.sum()
weights = fn_weighted_avg( kernel_pca.eigenvalues_ )
reconstructed_NDAQ_index = np.dot( transformed_pca_5, weights )
pd.DataFrame( reconstructed_NDAQ_index )  Those results from transform() are normalized using the weights indicated by the eigenvalues, computed with dot matrix multiplication. We then create a copy of the Dow time series pandas DataFrame with the copy() method, and combine it with the reconstructed values in the df_combined DataFrame.
Those results from transform() are normalized using the weights indicated by the eigenvalues, computed with dot matrix multiplication. We then create a copy of the Dow time series pandas DataFrame with the copy() method, and combine it with the reconstructed values in the df_combined DataFrame.
# Combine DJIA and PCA index for comparison
df_combined = djia_2017.copy()
df_combined['pca_5'] = reconstructed_NDAQ_index
df_combinedThe new DataFrame is normalized by z-scoring, and plotted out to see how well the reconstructed PCA index tracks the original Dow movements. This gives us the following output:
# standardization
df_combined = df_combined.apply(fn_z_score)
df_combined.plot( figsize=(12, 8) )![]() from quandl
from quandl from yfinance(more complete and accurate)
from yfinance(more complete and accurate)
The preceding graph shows the original Dow index against the reconstructed Dow index with five principal components for the year 2017.
Stationary and non-stationary time series
It is important that time series data that's used for statistical analysis is stationary in order to perform statistical modeling correctly, as such usages may be for prediction and forecasting. This section introduces the concepts of stationarity and non-stationarity in time series data.
Stationarity and non-stationarity
In empirical time series studies, price movements are observed to drift toward some long-term mean, either upwards or downwards.
- A stationary time series is one whose statistical properties, such as mean, variance, and autocorrelation, are constant over time.
 The daily change in the Google closing stock price has no trend, seasonality or cyclic behaviour(In general, the average length of cycles is longer than the length of a seasonal pattern, and the magnitudes of cycles tend to be more variable than the magnitudes of seasonal patterns.). There are random fluctuations which do not appear to be very predictable, and no strong patterns that would help with developing a forecasting model.
The daily change in the Google closing stock price has no trend, seasonality or cyclic behaviour(In general, the average length of cycles is longer than the length of a seasonal pattern, and the magnitudes of cycles tend to be more variable than the magnitudes of seasonal patterns.). There are random fluctuations which do not appear to be very predictable, and no strong patterns that would help with developing a forecasting model. - Conversely, observations on non-stationary time series data have their statistical properties change over time, mostly likely due to
- trends,
 (The US treasury bill contracts (top right) show results from the Chicago market for 100 consecutive trading days in 1981. Here there is no seasonality, but an obvious downward trend. Possibly, if we had a much longer series, we would see that this downward trend is actually part of a long cycle, but when viewed over only 100 days it appears to be a trend.)
(The US treasury bill contracts (top right) show results from the Chicago market for 100 consecutive trading days in 1981. Here there is no seasonality, but an obvious downward trend. Possibly, if we had a much longer series, we would see that this downward trend is actually part of a long cycle, but when viewed over only 100 days it appears to be a trend.)
A trend exists when there is a long-term increase or decrease in the data. It does not have to be linear. Sometimes we will refer to a trend as “changing direction”, when it might go from an increasing trend to a decreasing trend. There is a trend in the anti-diabetic抗糖尿病药 drug sales data shown in Figure 2.2.
- seasonality,(A seasonal pattern occurs when a time series is affected by seasonal factors such as the time of the year or the day of the week. Seasonality is always of a fixed and known frequency. The monthly sales of antidiabetic drugs above shows seasonality which is induced partly by the change in the cost of the drugs at the end of the calendar year.
) - presence of a unit root,
- or a combination of all three.
- trends,
In time series analysis, it is assumed that the data of the underlying process is stationary. Otherwise, modeling from non-stationary data may produce unpredictable results. This would lead to a condition known as spurious regression虚假回归. Spurious/ˈspjʊriəs/ regression is a regression that produces misleading statistical evidence of relationships between independent non-stationary variables. In order to receive consistent and reliable results, non-stationary data needs to be transformed into stationary data.
Checking for stationarity
There are a number of ways to check whether time series data is stationary or non-stationary:
- Through visualizations: You can review a time series graph for obvious indication of trends or seasonality.
- Through statistical summaries: You can review the statistical summaries of your data significant differences. For example, you can partition your time series data and compare the mean and variance of each group.
- Through statistical tests: You can use statistical tests such as the Augmented Dickey-Fuller Test to check if stationarity expectations have been met or violated.
Types of non-stationary processes
The following points help to identify non-stationary behavior in time series data for consideration in transforming stationary data:
- Pure random walk: A process with a unit root or a stochastic trend. It is a non-mean reverting process with a variance that evolves over time and goes to infinity.
- Random walk with drift: A process with a random walk and a constant drift.
- Deterministic trend: A process with a mean that grows around a fixed trend, which is constant and independent of time.
- Random walk with drift and deterministic trend: A process combining a random walk with a drift component, and a deterministic trend.
Types of stationary processes
These are a number of definitions of stationarity that you may come across in time series studies:
- Stationary process: A process that generates a stationary series of observations.
- Trend stationary: A process that does not exhibit a trend.
- Seasonal stationary: A process that does not exhibit seasonality.
- Strictly stationary: Also known as strongly stationary. A process whose unconditional joint probability distribution of random variables does not change when shifted in time (or along the x axis).
- Weakly stationary: Also known as covariance-stationary, or second-order stationary. A process whose mean, variance, and correlation of random variables doesn't change when shifted in time.
The Augmented Dickey-Fuller Test
An Augmented Dickey-Fuller Test (ADF) is a type of statistical test that determines whether a unit root is present in time series data. Unit roots can cause unpredictable results in time series analysis. A null hypothesis is formed on the unit root test to determine how strongly time series data is affected by a trend.
- By accepting the null hypothesis, we accept the evidence that the time series data is non-stationary.
- By rejecting the null hypothesis, or accepting the alternative hypothesis, we accept the evidence that the time series data is generated by a stationary process. This process is also known as trend-stationary.
- Values of the ADF test statistic are negative. Lower values of ADF indicates stronger rejection of the null hypothesis. the stronger the rejection of the hypothesis that there is a unit root at some level of confidence.
- The testing procedure for the ADF test is the same as for the Dickey–Fuller test but it is applied to the model https://blog.csdn.net/Linli522362242/article/details/121721868


Here are some basic autoregression models for use in ADF testing:
Here, ![]() is the drift constant,
is the drift constant, ![]() is the coefficient on a time trend,
is the coefficient on a time trend, ![]() is the coefficient of our hypothesis,
is the coefficient of our hypothesis, ![]() is the lag order of the first-differences autoregressive process
is the lag order of the first-differences autoregressive process
(![]()
where![]() are the parameters of the model,
are the parameters of the model, ![]() is a constant, and
is a constant, and ![]() is white noise(lagged value). This can be equivalently written using the backshift operator B后移运算符 as
is white noise(lagged value). This can be equivalently written using the backshift operator B后移运算符 as so that, moving the summation term to the left side and using polynomial notation, we have
so that, moving the summation term to the left side and using polynomial notation, we have![]()
Some parameter constraints are necessary for the model to remain wide-sense stationary保持广义平稳. For example, processes in the AR(1) model with![]() are not stationary. More generally, for an AR(p) model to be wide-sense stationary, the roots of the polynomial ORmust lie outside the unit circle, i.e., each (complex) root must satisfy .
are not stationary. More generally, for an AR(p) model to be wide-sense stationary, the roots of the polynomial ORmust lie outside the unit circle, i.e., each (complex) root must satisfy .
![]() OR
OR![]() must lie outside the unit circle
must lie outside the unit circle , i.e., each (complex) root
, i.e., each (complex) root![]() must satisfy
must satisfy ![]() .
.
), and ![]() is an independent and identically distributed residual term. When
is an independent and identically distributed residual term. When ![]() and
and ![]() , the model is a random walk process. When
, the model is a random walk process. When ![]() , the model is a random walk with a drift process. The length of the lag
, the model is a random walk with a drift process. The length of the lag ![]() is to be chosen so that the residuals are not serially correlated (By including lags of the order
is to be chosen so that the residuals are not serially correlated (By including lags of the order ![]() the ADF formulation allows for higher-order autoregressive processes. This means that the lag length p has to be determined when applying the test. One possible approach is to test down from high orders and examine the t-values on coefficients). An alternative approach is to examine information criteria for choosing lags are by minimizing the Akaike information criterion (AIC
the ADF formulation allows for higher-order autoregressive processes. This means that the lag length p has to be determined when applying the test. One possible approach is to test down from high orders and examine the t-values on coefficients). An alternative approach is to examine information criteria for choosing lags are by minimizing the Akaike information criterion (AIC![]() ), the Bayesian information criterion (BIC
), the Bayesian information criterion (BIC ![]() ), and the Hannan-Quinn information criterion.
), and the Hannan-Quinn information criterion.
- n is the number of instances, the number of data points in X, the number of observations, or equivalently, the sample size;
- k is the number of parameters learned by the model, the number of parameters estimated by the model. For example, in multiple linear regression, the estimated parameters are the intercept, the
 slope parameters, and the constant variance of the errors; thus
slope parameters, and the constant variance of the errors; thus  ,
,  is the maximized value of the likelihood function of the model M. i.e.
is the maximized value of the likelihood function of the model M. i.e.  , where
, where  are the parameter values that maximize the likelihood function,
are the parameter values that maximize the likelihood function,  = the observed data;
= the observed data;- Both the BIC and the AIC penalize models that have more parameters to learn (e.g., more clusters) and reward models that fit the data well. They often end up selecting the same model. When they differ, the model selected by the BIC tends to be simpler(fewer parameters, 考虑了样本数量,样本数量过多时,可有效防止模型精度过高造成的模型复杂度过高) than the one selected by the AIC, but tends to not fit the data quite as well (this is especially true for larger datasets).
Likelihood Function
The terms “probability” and “likelihood” are often used interchangeably in the English language, but they have very different meanings in statistics.
- Given a statistical model with some parameters θ, the word “probability” is used to describe how plausible/ ˈplɔːzəb(ə)l /可信的,可靠的 a future outcome x is (knowing the parameter values θ)“概率”一词用于描述未来结果 x 的可信度,
- while the word “likelihood” is used to describe how plausible a particular set of parameter values θ are, after the outcome x is known.
![]() Figure 9-20. A model’s parametric function (top left), and some derived functions: a PDF(Probability Density Function, lower left, 概率密度函数PDF是对连续 随机变量定义的,本⾝不是概率,只有对连续 随机变量的概率密度函数PDF在某区间内进⾏积分后才是概率), a likelihood function (top right), and a log likelihood function (lower right)
Figure 9-20. A model’s parametric function (top left), and some derived functions: a PDF(Probability Density Function, lower left, 概率密度函数PDF是对连续 随机变量定义的,本⾝不是概率,只有对连续 随机变量的概率密度函数PDF在某区间内进⾏积分后才是概率), a likelihood function (top right), and a log likelihood function (lower right)
Consider a 1D mixture model of two Gaussian distributions centered at –4 and +1. For simplicity, this toy model has a single parameter θ that controls the standard deviations of both distributions. The top-left contour plot等高线图 in Figure 9-20 shows the entire model f(x; θ) as a function of both x and θ.
- To estimate the probability distribution of a future outcome x, you need to set the model parameter θ. For example, if you set θ to 1.3 (the horizontal line), you get the probability density function f(x; θ=1.3) shown in the lower-left plot. Say you want to estimate the probability that x will fall between –2 and +2. You must calculate the integral of the PDF on this range (i.e., the surface of the shaded region).
- But what if you don’t know θ, and instead if you have observed a single instance x=2.5 (the vertical line in the upper-left plot)? In this case, you get the likelihood function ℒ(θ|x=2.5)=f(x=2.5; θ), represented in the upper-right plot.https://blog.csdn.net/Linli522362242/article/details/96480059
In short, the PDF is a function of x (with θ fixed), while the likelihood function is a function of θ (with x fixed). It is important to understand that the likelihood function is not a probability distribution:
- if you integrate a probability distribution over all possible values of x, you always get 1;
- but if you integrate the likelihood function over all possible values of θ, the result can be any positive value.
Given a dataset X, a common task is to try to estimate the most likely values for the model parameters. To do this, you must find the values that maximize the likelihood function, given X. In this example, if you have observed a single instance x=2.5, the maximum likelihood estimate (MLE) of θ is ![]() . If a prior probability distribution g over θ exists 如果存在关于 θ 的先验概率分布 g, it is possible to take it into account by maximizing ℒ(θ|x)g(θ) rather than just maximizing ℒ(θ|x). This is called maximum a-posteriori (MAP) estimation最大后验 (MAP) 估计. Since MAP constrains the parameter values, you can think of it as a regularized version of MLE.
. If a prior probability distribution g over θ exists 如果存在关于 θ 的先验概率分布 g, it is possible to take it into account by maximizing ℒ(θ|x)g(θ) rather than just maximizing ℒ(θ|x). This is called maximum a-posteriori (MAP) estimation最大后验 (MAP) 估计. Since MAP constrains the parameter values, you can think of it as a regularized version of MLE.
Notice that maximizing the likelihood function is equivalent to maximizing its logarithm (represented in the lower-right hand plot in Figure 9-20). Indeed the logarithm is a strictly increasing function, so if θ maximizes the log likelihood, it also maximizes the likelihood. It turns out that it is generally easier to maximize the log likelihood. For example, if you observed several independent instances x(1) to x(m), you would need to find the value of θ that maximizes the product of the individual likelihood functions. But it is equivalent, and much simpler, to maximize the sum (not the product) of the log likelihood functions, thanks to the magic of the logarithm which converts products into sums: log(ab)=log(a)+log(b).
Once you have estimated ![]() , the value of θ that maximizes the likelihood function, then you are ready to compute
, the value of θ that maximizes the likelihood function, then you are ready to compute ![]() , which is the value used to compute the AIC and BIC; you can think of it as a measure of how well the model fits the data.
, which is the value used to compute the AIC and BIC; you can think of it as a measure of how well the model fits the data.
AIC和BIC主要用于模型的选择,AIC、BIC越小越好。
- 在对不同模型进行比较时,AIC、BIC降低越多,说明该模型的拟合效果越好;选择最优模型的指导思想是从两个方面去考察:一个是似然函数最大化,另一个是模型中的未知参数个数最小化。似然函数值越大说明模型拟合的效果越好,但是我们不能单纯地以拟合精度来衡量模型的优劣,这样回导致模型中未知参数k越来越多,模型变得越来越复杂,会造成过拟合。所以一个好的模型应该是拟合精度和未知参数个数的综合最优化配置。
- 当两个模型之间存在较大差异时,差异主要体现在似然函数项,当似然函数差异不显著时,上式第一项,即模型复杂度则起作用,从而参数个数少的模型是较好的选择。
- AIC: 一般而言,当模型复杂度提高(k增大)时,似然函数
 也会增大,从而使AIC变小,但是k过大时,似然函数增速减缓,导致AIC增大,模型过于复杂容易造成过拟合现象。目标是选取AIC最小的模型,AIC不仅要提高模型拟合度(极大似然),而且引入了惩罚项,使模型参数尽可能少,有助于降低过拟合的可能性。
也会增大,从而使AIC变小,但是k过大时,似然函数增速减缓,导致AIC增大,模型过于复杂容易造成过拟合现象。目标是选取AIC最小的模型,AIC不仅要提高模型拟合度(极大似然),而且引入了惩罚项,使模型参数尽可能少,有助于降低过拟合的可能性。 - AIC和BIC均引入了与模型参数个数相关的惩罚项,BIC的惩罚项比AIC的大,考虑了样本数量,样本数量过多时,可有效防止模型精度过高造成的模型复杂度过高(kln(n)惩罚项在维数过大且训练样本数据相对较少的情况下,可以有效避免出现维度灾难现象。)
Both BIC and AIC penalize models that have more parameters to learn (e.g., more clusters), and reward models that fit the data well (i.e., models that give a high likelihood to the observed data).https://blog.csdn.net/Linli522362242/article/details/105973507
statsmodels.tsa.seasonal.seasonal_decompose — statsmodels
This is a naive decomposition. More sophisticated methods should be preferred.
The additive model is Y[t] = T[t] + S[t] + e[t]
The multiplicative model is Y[t] = T[t] * S[t] * e[t]
The results are obtained by first estimating the trend by applying a convolution filter to the data. The trend is then removed from the series and the average of this de-trended series for each period is the returned seasonal component.
![]()
![]() https://blog.csdn.net/Linli522362242/article/details/121721868
https://blog.csdn.net/Linli522362242/article/details/121721868
Here, ![]() is the drift constant,
is the drift constant, ![]() is the coefficient on a time trend,
is the coefficient on a time trend, ![]() is the coefficient of our hypothesis,
is the coefficient of our hypothesis, ![]() is the lag order of the first-differences autoregressive process, and
is the lag order of the first-differences autoregressive process, and ![]() is an independent and identically distributed residual term. When
is an independent and identically distributed residual term. When ![]() and
and ![]() , the model is a random walk process. When
, the model is a random walk process. When ![]() , the model is a random walk with a drift process. The length of the lag
, the model is a random walk with a drift process. The length of the lag ![]() is to be chosen so that the residuals are not serially correlated. Some approaches for examining the information criteria for choosing lags are by minimizing the Akaike information criterion (AIC
is to be chosen so that the residuals are not serially correlated. Some approaches for examining the information criteria for choosing lags are by minimizing the Akaike information criterion (AIC![]() ), the Bayesian information criterion (BIC
), the Bayesian information criterion (BIC ![]() ), and the Hannan-Quinn information criterion.
), and the Hannan-Quinn information criterion.
The hypothesis can then be formulated as follows:
- Null hypothesis,
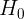 : If failed to be rejected, it suggests that the time series contains a unit root and is non-stationary
: If failed to be rejected, it suggests that the time series contains a unit root and is non-stationary
A null hypothesis is formed on the unit root test to determine how strongly time series data is affected by a trend. - Alternate hypothesis,
 or
or  : If
: If 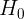 is rejected, it suggests that the time series does not contain a unit root and is stationary
is rejected, it suggests that the time series does not contain a unit root and is stationary
To accept or reject the null hypothesis![]() , we use the p-value. We reject the null hypothesis
, we use the p-value. We reject the null hypothesis![]() if the p-value falls below a threshold value such as 5% or even 1%. We can fail to reject the null hypothesis
if the p-value falls below a threshold value such as 5% or even 1%. We can fail to reject the null hypothesis![]() if the p-value is above this threshold value and consider the time series as non-stationary. In other words, if our threshold value is 5%, or 0.05, note the following:
if the p-value is above this threshold value and consider the time series as non-stationary. In other words, if our threshold value is 5%, or 0.05, note the following:
- p-value > 0.05: We fail to reject the null hypothesis
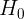 and conclude that the data has a unit root and is non-stationary
and conclude that the data has a unit root and is non-stationary - p-value ≤ 0.05: We reject the null hypothesis
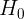 and conclude that the data does not contain a unit root and is stationary
and conclude that the data does not contain a unit root and is stationary
The statsmodels library provides the adfuller() function that implements this test.
Analyzing a time series with trends
Let's examine a time series dataset. Take, for example, the prices of gold futures traded on the CME(Chicago Mercantile Exchange). On Quandl, the gold futures continuous contract is available for download with the following code: CHRIS/CME_GC1 . This data is curated/ ˈkjʊərət/操持 by the Wiki Continuous Futures community group, taking into account the front month contracts only. The sixth column of the dataset contains the settlement prices结算价格. The following code downloads the dataset from the year 2000 onward:
import quandl
QUANDL_API_KEY = 'sKqHwnHr8rNWK-3s5imS'
quandl.ApiConfig.api_key = QUANDL_API_KEY
df_full = quandl.get( 'CHRIS/CME_GC1',
collapse='monthly',
start_date='2000-01-01'
)
df_fulldf = pd.DataFrame(df_full['Settle'])
dfCompute the rolling mean and standard deviation into the df_mean and df_std variables, respectively, with a window period of one year:
sample frequency
B business day frequency
C custom business day frequency (experimental)
D calendar day frequency
W weekly frequency
M month end frequency
SM semi-month end frequency (15th and end of month)
BM business month end frequency
CBM custom business month end frequency
MS month start frequency
SMS semi-month start frequency (1st and 15th)
BMS business month start frequency
CBMS custom business month start frequency
Q quarter end frequency
BQ business quarter endfrequency
QS quarter start frequency
BQS business quarter start frequency
A year end frequency
BA, BY business year end frequency
AS, YS year start frequency
BAS, BYS business year start frequency
BH business hour frequency
H hourly frequency
T, min minutely frequency
S secondly frequency
L, ms milliseconds
U, us microseconds
N nanoseconds# 'MS' : month start frequency
df_settle = df['Settle'].resample('MS').ffill().dropna()
pd.DataFrame( df_settle )The resample() method helps to ensure that the data is smoothed out on a monthly basis, and the ffill() method forward fills any missing values.
A list of useful common time series frequencies for specifying the resample() method can be found at https://pandas.pydata.org/docs/user_guide/timeseries.html.![]()
# DataFrame.rolling(window, min_periods=None, center=False, win_type=None, on=None, axis=0, closed=None, method='single')
df_rolling = df_settle.rolling(12)
df_mean = df_rolling.mean()
df_meandf_std = df_rolling.std()
df_stdLet's visualize the plot of the rolling mean against the original time series:
plt.figure( figsize=(12,8) )
plt.plot( df_settle, label='Original' )
plt.plot( df_mean, label='Mean' )
plt.legend()
the mean exhibiting an overall upward trend.
Visualizing the rolling standard deviation separately, we get the following:
df_std.plot( figsize=(12,8) )Using the statsmodels module, perform an ADF unit root test on our dataset with the adfuller() method:
from statsmodels.tsa.stattools import adfuller
result = adfuller( df_settle )
resultThe adfuller() method returns a tuple of seven values. Particularly, we are interested in the first, second, and fifth values, which give us the test statistic, p-value , and a dictionary of critical values, respectively.
(-0.6727450369450463, |
The test statistic. |
0.8536834573174014, |
MacKinnon’s approximate p-value based on MacKinnon |
11,11 = rolling_window-1 = (12-1) |
The number of lags used. |
245, |
The number of observations used for the ADF regression and calculation of the critical values. 245=257 -12 |
{'1%': -3.4573260719088132, |
Critical values for the test statistic at the 1 % |
'5%': -2.873410402808354, |
Critical values for the test statistic at the 5 % |
'10%': -2.573095980841316}, |
Critical values for the test statistic at the 10 % |
2608.4357297508386) |
The maximized information criterion if autolag is not None.(default autolag='AIC', ) |
print( 'ADF test statistic.: ', result[0] )
# MacKinnon’s approximate p-value based on MacKinnon
print( 'p-value:', result[1] )
critical_values = result[4]
for key, value in critical_values.items():
print( 'Critical value (%s): %.3f' % (key, value) ) Observe from the plots that the mean and standard deviations swing over time, with the mean exhibiting an overall upward trend. The ADF test statistic value is more than the critical values (especially at 5%), and the p-value is more than 0.05. With these, we cannot reject the null hypothesis![]() that there is a unit root and consider that our data is non-stationary.
that there is a unit root and consider that our data is non-stationary.
Making a time series stationary
A non-stationary time series data is likely to be affected by a trend or seasonality. Trending time series data has a mean that is not constant over time. Data that is affected by seasonality have variations at specific intervals in time. In making a time series data stationary, the trend and seasonality effects have to be removed. Detrending, differencing, and decomposition are such methods. The resulting stationary data is then suitable for statistical forecasting.
Let's look at all three methods in detail.
Detrending
The process of removing a trend line from a non-stationary data is known as detrending. This involves a transformation step that normalizes large values into smaller ones. Examples could be a logarithmic function, a square root function, or even a cube root. A further step is to subtract the transformation from the moving average.
Let's perform detrending on the same dataset, df_settle , with logarithmic transformation and subtracting from the moving average of two periods, as given in the following Python code:
import numpy as np
# 'MS' : month start frequency
# df_settle = df['Settle'].resample('MS').ffill().dropna()
df_log = np.log( df_settle )
df_logThe df_log variable is our transformed pandas DataFrame by logarithmic function using the numpy module, and the df_detrend variable contains the detrended data.
df_log_ma = df_log.rolling(2).mean()
df_detrend = df_log - df_log_ma
df_detrend.dropna( inplace=True )
df_detrend5.684260-(5.646153 + 5.684260)/2 = 0.019053500000000057 ==> back_rolling
# Mean and standard deviation of detrended data
df_detrend_rolling = df_detrend.rolling(12)
df_detrend_ma = df_detrend_rolling.mean()
df_detrend_std = df_detrend_rolling.std()
# Plot
plt.figure(figsize=(12, 8))
plt.plot(df_detrend, label='Detrended', c='k')
plt.plot(df_detrend_ma, label='mean', c='y')
plt.plot(df_detrend_std, label='std', c='b')
plt.legend(loc='upper right');We plot this detrended data to visualize its mean and standard deviation over a rolling one-year period.
Observe that the mean and standard deviation do not exhibit a long-term trend.
Looking at the ADF test statistic for the detrended data, we get the following:
from statsmodels.tsa.stattools import adfuller
result = adfuller( df_detrend )
print('ADF test statistic: ', result[0])
print('p-value: %.5f' % result[1])
critical_values = result[4]
for key, value in critical_values.items():
print( 'Critical value (%s): %.3f' % (key, value) ) The p-value for this detrended data is less than 0.05. Our ADF test statistic is lower than all the critical values. We can reject the null hypothesis
The p-value for this detrended data is less than 0.05. Our ADF test statistic is lower than all the critical values. We can reject the null hypothesis![]() and say that this data is stationary.
and say that this data is stationary.
result (-17.83616011271889 |
The test statistic. |
3.119281798723769e-30 |
MacKinnon’s approximate p-value based on MacKinnon |
0 |
The number of lags used. |
255, |
The number of observations used for the ADF regression and calculation of the critical values. |
{'1%': -3.4562572510874396, |
Critical values for the test statistic at the 1 % |
'5%': -2.8729420379793598 |
Critical values for the test statistic at the 5 % |
'10%': -2.5728461399461744}, |
Critical values for the test statistic at the 10 % |
-1104.2171260985874) |
The maximized information criterion if autolag is not None.(default autolag='AIC', ) |
result = adfuller( df_detrend, autolag='BIC' )
result######################https://blog.csdn.net/Linli522362242/article/details/121406833
Removing trend by differencing
Log returns(log_return): between two times 0 < s < t are normally distributed.
between two times 0 < s < t are normally distributed.
df_settle![]()
df_detrend=np.log( df_settle / df_settle.shift(1) ).dropna()
df_detrendnp.log( 294.2/283.2 ) = 0.03810644634777238 ==>shift forward
![]()
# Mean and standard deviation of detrended data
df_detrend_rolling = df_detrend.rolling(12)
df_detrend_ma = df_detrend_rolling.mean()
df_detrend_std = df_detrend_rolling.std()
# Plot
plt.figure(figsize=(12, 8))
plt.plot(df_detrend, label='Detrended', c='k')
plt.plot(df_detrend_ma, label='mean', c='y')
plt.plot(df_detrend_std, label='std', c='b')
plt.legend(loc='upper right');from statsmodels.tsa.stattools import adfuller
result = adfuller( df_detrend )
print('ADF test statistic: ', result[0])
print('p-value: %.5f' % result[1])
critical_values = result[4]
for key, value in critical_values.items():
print( 'Critical value (%s): %.3f' % (key, value) ) (-17.836160112718915, |
The test statistic. |
3.11928179872368e-30, |
MacKinnon’s approximate p-value based on MacKinnon |
0 |
The number of lags used. |
255, |
The number of observations used for the ADF regression and calculation of the critical values. |
{'1%': -3.4562572510874396, |
Critical values for the test statistic at the 1 % |
'5%': -2.8729420379793598 |
Critical values for the test statistic at the 5 % |
'10%': -2.5728461399461744}, |
Critical values for the test statistic at the 10 % |
-772.8927737909331) |
The maximized information criterion if autolag is not None.(default autolag='AIC', ) |
result = adfuller( df_detrend, autolag='BIC' )
result######################
Removing trend by differencing
Differencing involves the difference of time series values with a time lag. The first-order difference of the time series is given by the following formula:![]()
Log returns(log_return): between two times 0 < s < t are normally distributed.
between two times 0 < s < t are normally distributed.
We can reuse the df_log variable in the previous section as our logarithmic transformed time series, and utilize the diff() and shift() methods of NumPy modules in our differencing, with the following code:
df_log_diff = df_log.diff( periods=3 ).dropna()
df_log_diffshift(3) : shift forward 3 periods
![]()
# Mean and standard deviation of differenced data
df_diff_rolling = df_log_diff.rolling(12)
df_diff_ma = df_diff_rolling.mean()
df_diff_ma # ma : moving averagedf_diff_std = df_diff_rolling.std()
df_diff_std# Plot the stationary data
plt.figure( figsize=(12,8) )
plt.plot( df_log_diff, label='Differenced', c='k' )
plt.plot( df_diff_ma, label='mean', c='y' )
plt.plot( df_diff_std, label='std', c='b' )
plt.legend( loc='upper right' ) 
Observe from the plots that the rolling mean and standard deviation tend to change very little over time.
Looking at our ADF test statistic, we get the following:
from statsmodels.tsa.stattools import adfuller
result = adfuller( df_log_diff )
print( 'ADF test statistic:', result[0] )
print( 'p-value: %.5f' % result[1] )
critical_values = result[4]
for key, value in critical_values.items():
print( 'Critical value (%s): %.3f' % (key, value) )result -3.4008135526634544 |
The test statistic. |
0.010931168900429722, |
MacKinnon’s approximate p-value based on MacKinnon |
12, |
The number of lags used. |
241, |
The number of observations used for the ADF regression and calculation of the critical values. |
{'1%': -3.4577787098622674,
|
Critical values for the test statistic at the 1 % |
'5%': -2.873608704758507, |
Critical values for the test statistic at the 5 % |
'10%': -2.573201765981991}, |
Critical values for the test statistic at the 10 % |
-727.8273552142234) |
The maximized information criterion if autolag is not None.(default autolag='AIC', ) |
result = adfuller( df_log_diff , autolag='BIC')
result (-5.503687089499893, |
The test statistic. |
2.044329397896393e-06, |
MacKinnon’s approximate p-value based on MacKinnon |
3, |
The number of lags used. |
250, |
The number of observations used for the ADF regression and calculation of the critical values. |
{'1%': -3.456780859712, |
Critical values for the test statistic at the 1 % |
'5%': -2.8731715065600003, |
Critical values for the test statistic at the 5 % |
'10%': -2.572968544}, |
Critical values for the test statistic at the 10 % |
-689.0343765535242) |
The maximized information criterion if autolag is not None.(default autolag='BIC', ) |
From the ADF test, the p-value for this data is less than 0.05. Our ADF test statistic is lower than the 5% critical value, indicating a 95% confidence level that this data is stationary. We can reject the null hypothesis![]() and say that this data is stationary.
and say that this data is stationary.
Seasonal decomposing
Decomposing involves modeling both the trend and seasonality, and then removing them. We can use the statsmodel.tsa.seasonal module to model a non-stationary time series dataset using moving averages and remove its trend and seasonal components.
By reusing our df_log variable containing the logarithm of our dataset from the previous section, we get the following:
two_sided bool, optional
The moving average method used in filtering. If True (default), a centered moving average is computed using the filt. If False, the filter coefficients are for past values only.
from statsmodels.tsa.seasonal import seasonal_decompose
# Seasonal decomposition using moving averages.
# df_log = np.log( df_settle )
# statsmodels.tsa.seasonal.seasonal_decompose(x, model='additive',
decompose_result = seasonal_decompose( df_log.dropna(), period = 12 )
df_trend = decompose_result.trend
df_season = decompose_result.seasonal
df_residual = decompose_result.residThe seasonal_decompose() method of statsmodels.tsa.seasonal requires a parameter, period, which is an integer value specifying the number of periods per seasonal cycle. Since we are using monthly data, we expect 12 periods in a seasonal year. The method returns an object with three attributes, mainly the trend and seasonal components, as well as the final pandas series data with its trend and seasonal components removed.
More information on the seasonal_decompose() method of the statsmodels.tsa.seasonal module can be found at https://www.statsmodels.org/dev/generated/statsmodels.tsa.seasonal.seasonal_decompose.html
Let's visualize the different plots by running the following Python code:
# https://matplotlib.org/stable/tutorials/introductory/customizing.html
plt.style.use('seaborn-white')
plt.rcParams['figure.figsize'] = (10,8)
#plt.rcdefaults()
fig = decompose_result.plot()
# pd.DataFrame( decompose_result.resid ).plot(figsize=(10,3))
fig.tight_layout()
plt.show()if you like the draw Resid plot with line:
fig = decompose_result.plot()
axs = fig.get_axes()
axs[3].clear()
axs[3].plot(decompose_result.resid)df_residual_diff = df_residual.diff().dropna()
# Mean and standard deviation of differenced data
df_residual_diff_rolling = df_residual_diff.rolling(12)
df_residual_diff_rolling_ma = df_residual_diff_rolling.mean()
df_residual_diff_rolling_std = df_residual_diff_rolling.std()
# Plot the stationary data
plt.figure( figsize=(12,8) )
plt.plot( df_residual_diff, label='Differenced', c='k' )
plt.plot( df_residual_diff_rolling_ma, label='Mean', c='b' )
plt.plot( df_residual_diff_rolling_std, label='Std', c='y' )
plt.legend()Observe from the plots that the rolling mean and standard deviation tend to change very little over time.
By checking our residual data for stationarity, we get the following:
from statsmodels.tsa.stattools import adfuller
result = adfuller( df_residual.dropna() )
result (-6.851743406274058, |
The test statistic. |
1.6874183653005745e-09, |
MacKinnon’s approximate p-value based on MacKinnon |
12, |
The number of lags used. |
232, |
The number of observations used for the ADF regression and calculation of the critical values. |
{'1%': -3.458854867412691, |
Critical values for the test statistic at the 1 % |
'5%': -2.8740800599399323, |
Critical values for the test statistic at the 5 % |
'10%': -2.573453223097503}, |
Critical values for the test statistic at the 10 % |
-854.884067978267) |
The maximized information criterion if autolag is not None.(default autolag='AIC', ) |
result = adfuller( df_residual.dropna(), autolag='BIC' )
result (-9.674116444744383, |
The test statistic. |
1.2540370602752483e-16, |
MacKinnon’s approximate p-value based on MacKinnon |
0, |
The number of lags used. |
244, |
The number of observations used for the ADF regression and calculation of the critical values. |
{'1%': -3.457437824930831, |
Critical values for the test statistic at the 1 % |
'5%': -2.873459364726563, |
Critical values for the test statistic at the 5 % |
'10%': -2.573122099570008}, |
Critical values for the test statistic at the 10 % |
-827.6316414544348) |
The maximized information criterion if autolag is not None.(default autolag='BIC', |
From the ADF test, the p-value for this data is less than 0.05. Our ADF test statistic is lower than all the critical values. We can reject the null hypothesis![]() and say that this data is stationary.
and say that this data is stationary.
Manual decomposition-same with seasonal_decompose & convolution_filter
![]()
statsmodels.tsa.filters.filtertools.convolution_filter — statsmodels
statsmodels.tsa.filters.filtertools.convolution_filter(x, filt, nsides=2)
https://blog.csdn.net/Linli522362242/article/details/108414534
![]() ==>
==> ![]()
In nsides == 1, x is filtered
y[n] = filt[0]*x[n-1] + ... + filt[n_filt-1]*x[n-n_filt]where n_filt is len(filt).
If nsides == 2, x is filtered around lag 0
y[n] = filt[0]*x[n - n_filt/2] + ... + filt[n_filt / 2] * x[n]
+ ... + x[n + n_filt/2]where n_filt is len(filt). If n_filt is even, then more of the filter is forward in time than backward.
If filt is 1d or (nlags,1) one lag polynomial is applied to all variables (columns of x). If filt is 2d, (nlags, nvars) each series is independently filtered with its own lag polynomial, uses loop over nvar. This is different than the usual 2d vs 2d convolution.
period=12
# [0.5] + broadcasting==> 11 elements + [0.5]
np.array([0.5] + [1]*(period - 1) + [0.5]) / period![]()
nsides = int(two_sided) + 1, (default nsides=True) ==2
from statsmodels.tsa.filters.filtertools import convolution_filter
from pandas.core.nanops import nanmean as pd_nanmean
# df_log = np.log( df_settle )
seasonal_df = pd.DataFrame( df_log.dropna() )
#################### calulate the trend component ####################
# moving average
# seasonal_df["trend"] = seasonal_df["Settle"].rolling(window=12, center=True).mean()
########## seasonal_decompose - trend ##########
period=12
two_sided = True # Default
# The moving average method used in filtering.
# If True (default), a centered moving average is computed using the filt.
# If False, the filter coefficients are for past values only.
nsides = int(two_sided) + 1
seasonal_df["trend"] = convolution_filter(seasonal_df,
# [0.5] + broadcasting==> 11 elements + [0.5]
np.array([0.5] + [1]*(period - 1) + [0.5]) / period,
nsides
)# Bt
# n_filt is len(filt)
# y[n] = filt[0]*x[n - n_filt/2] + ... + filt[n_filt / 2] * x[n]
# + ... + x[n + n_filt/2]
# detrend the series # - moving average
seasonal_df["detrended"] = seasonal_df["Settle"] - seasonal_df["trend"]
#################### calculate the seasonal component ####################
# seasonal_df.index = pd.to_datetime( seasonal_df.index )
# seasonal_df["month"] = seasonal_df.index.month
# seasonal_df["seasonality"] = seasonal_df.groupby("month")["detrended"].transform("mean")
########## seasonal_decompose - seasonality ##########
def seasonal_mean(x, period):
"""
Return means for each period in x. period is an int that gives the
number of periods per cycle. E.g., 12 for monthly. NaNs are ignored
in the mean.
"""
return np.array([ pd_nanmean( x[i::period], axis=0 )
for i in range(period)
])
period_averages = seasonal_mean(seasonal_df["detrended"], period)
period_averages -= np.mean( period_averages, axis=0 )
seasonal = np.tile( period_averages.T,
len(seasonal_df["detrended"]) // period + 1
).T[:len(seasonal_df["detrended"])]
seasonal_df["seasonality"] = seasonal
# get the residuals
seasonal_df["resid"] = seasonal_df["detrended"] - seasonal_df["seasonality"]
seasonal_df[:20]df_residual_diff = seasonal_df["resid"].diff().dropna()
# Mean and standard deviation of differenced data
df_residual_diff_rolling = df_residual_diff.rolling(12)
df_residual_diff_rolling_ma = df_residual_diff_rolling.mean()
df_residual_diff_rolling_std = df_residual_diff_rolling.std()
# Plot the stationary data
plt.figure( figsize=(12,8) )
plt.plot( df_residual_diff, label='Differenced', c='k' )
plt.plot( df_residual_diff_rolling_ma, label='Mean', c='b' )
plt.plot( df_residual_diff_rolling_std, label='Std', c='y' )
plt.legend()from statsmodels.tsa.stattools import adfuller
result = adfuller( seasonal_df["resid"].dropna() )
resultfrom statsmodels.tsa.seasonal import DecomposeResult
manual_decomposition = DecomposeResult(
seasonal=seasonal_df["seasonality"],
trend=seasonal_df["trend"],
resid=seasonal_df["resid"],
# decompose_result.observed
observed=seasonal_df["Settle"],
)
def add_second_decomp_plot(fig, res, legend):
axs = fig.get_axes()
comps = ["trend", "seasonal", "resid"]
for ax, comp in zip(axs[1:], comps):
series = getattr(res, comp)
if comp == "resid":
ax.plot(series, marker="o", linestyle="none")
else:
ax.plot(series)
if comp == "trend":
ax.legend(legend, frameon=False)
fig = decompose_result.plot()
add_second_decomp_plot( fig, manual_decomposition, ["statsmodels", "manual"] );![]()
Manual decomposition-little different with seasonal_decompose
# from statsmodels.tsa.filters.filtertools import convolution_filter
# from pandas.core.nanops import nanmean as pd_nanmean
# df_log = np.log( df_settle )
seasonal_df = pd.DataFrame( df_log.dropna() )
#################### calulate the trend component ####################
# moving average
seasonal_df["trend"] = seasonal_df["Settle"].rolling(window=12, center=True).mean()
########## seasonal_decompose - trend ##########
# period=12
# two_sided = True # Default
# # The moving average method used in filtering.
# # If True (default), a centered moving average is computed using the filt.
# # If False, the filter coefficients are for past values only.
# nsides = int(two_sided) + 1
# seasonal_df["trend"] = convolution_filter(seasonal_df,
# # [0.5] + broadcasting==> 11 elements + [0.5]
# np.array([0.5] + [1]*(period - 1) + [0.5]) / period,
# nsides
# )# Bt
# # n_filt is len(filt)
# # y[n] = filt[0]*x[n - n_filt/2] + ... + filt[n_filt / 2] * x[n]
# # + ... + x[n + n_filt/2]
# detrend the series # - moving average
seasonal_df["detrended"] = seasonal_df["Settle"] - seasonal_df["trend"]
#################### calculate the seasonal component ####################
seasonal_df.index = pd.to_datetime( seasonal_df.index )
seasonal_df["month"] = seasonal_df.index.month
seasonal_df["seasonality"] = seasonal_df.groupby("month")["detrended"].transform("mean")
########## seasonal_decompose - seasonality ##########
# def seasonal_mean(x, period):
# """
# Return means for each period in x. period is an int that gives the
# number of periods per cycle. E.g., 12 for monthly. NaNs are ignored
# in the mean.
# """
# return np.array([ pd_nanmean( x[i::period], axis=0 )
# for i in range(period)
# ])
# period_averages = seasonal_mean(seasonal_df["detrended"], period)
# period_averages -= np.mean( period_averages, axis=0 )
# seasonal = np.tile( period_averages.T,
# len(seasonal_df["detrended"]) // period + 1
# ).T[:len(seasonal_df["detrended"])]
# seasonal_df["seasonality"] = seasonal
# get the residuals
seasonal_df["resid"] = seasonal_df["detrended"] - seasonal_df["seasonality"]
seasonal_df[:20]df_residual_diff = seasonal_df["resid"].diff().dropna()
# Mean and standard deviation of differenced data
df_residual_diff_rolling = df_residual_diff.rolling(12)
df_residual_diff_rolling_ma = df_residual_diff_rolling.mean()
df_residual_diff_rolling_std = df_residual_diff_rolling.std()
# Plot the stationary data
plt.figure( figsize=(12,8) )
plt.plot( df_residual_diff, label='Differenced', c='k' )
plt.plot( df_residual_diff_rolling_ma, label='Mean', c='b' )
plt.plot( df_residual_diff_rolling_std, label='Std', c='y' )
plt.legend()from statsmodels.tsa.stattools import adfuller
result = adfuller( seasonal_df["resid"].dropna() )
resultfrom statsmodels.tsa.seasonal import DecomposeResult
manual_decomposition = DecomposeResult(
seasonal=seasonal_df["seasonality"],
trend=seasonal_df["trend"],
resid=seasonal_df["resid"],
# decompose_result.observed
observed=seasonal_df["Settle"],
)
def add_second_decomp_plot(fig, res, legend):
axs = fig.get_axes()
comps = ["trend", "seasonal", "resid"]
for ax, comp in zip(axs[1:], comps):
series = getattr(res, comp)
if comp == "resid":
ax.plot(series, marker="o", linestyle="none")
else:
ax.plot(series)
if comp == "trend":
ax.legend(legend, frameon=False)
fig = decompose_result.plot()
add_second_decomp_plot( fig, manual_decomposition, ["statsmodels", "manual"] );Drawbacks of ADF testing
Here are some considerations when using ADF tests for reliable checking of non-stationary data:
- The ADF test do not truly tell apart between pure and non-unit root generating processes. In long-term moving average processes, the ADF tests becomes biased偏差 in rejecting the null hypothesis. Other stationarity testing methods such as the Kwiatkowski–Phillips–Schmidt–Shin (KPSS) tests and the Phillips-Perron test take a different approach in treating the presence of unit roots.
- There is no fixed methodology in determining the lag length p.
- If p is too small, the remaining serial correlation in the errors may affect the size of the test.
- If p is too large, the power of the test will deteriorate/dɪˈtɪriəreɪt/恶化.
- Additional consideration is to be given for this lag order.
- As deterministic terms are added to the test regressions, the power of unit root tests diminishes.
Forecasting and predicting a time series
In the previous section, we identified non-stationarity in time series data and discussed techniques for making time series data stationary. With stationary data, we can proceed to perform statistical modeling such as prediction and forecasting. Prediction involves generating best estimates of in-sample data. Forecasting involves generating best estimates of out-of-sample data. Predicting future values is based on previously observed values. One such commonly used method is the Autoregressive Integrated Moving Average.
(ARIMA) About the Autoregressive Integrated Moving Average
The Autoregressive Integrated Moving Average (ARIMA) is a forecasting model for stationary time series based on linear regression.(ARIMA models aim to describe the autocorrelations in the data.) As its name suggests, it is based on three components:
- Autoregression (AR): A model that uses the dependency between an observation and its lagged values
- Integrated (I): The use of differencing an observation with an observation from a previous time stamp in making the time series stationary
- Moving average (MA): A model that uses the dependency between an observed error term and a combination of previous error terms,

 OR
OR  OR
OR 
 are the parameters of the model,
are the parameters of the model,  is a constant, and
is a constant, and is white noise(lagged value).
is white noise(lagged value).
ARIMA models are referenced by the notation ARIMA(p, d, q), which corresponds to the parameters of the three components. Non-seasonal ARIMA models can be specified by changing the values of p, d, and q, as follows:
- ARIMA(p,0,0):
First-order autoregressive model, notated by AR(p).
p is the lag order, indicating the number of lagged observations in the model.
AR(1) :
 or
or 
For example, ARIMA(2,0,0) is AR(2) and represented as follows:

Here, and
and  are parameters for the model. and
are parameters for the model. and  is white noise
is white noise
In a multiple regression model, we forecast the variable of interest using a linear combination of predictors. In an autoregression model, we forecast the variable of interest using a linear combination of past values of the variable
of interest using a linear combination of past values of the variable . The term autoregression indicates that it is a regression of the variable against itself.
. The term autoregression indicates that it is a regression of the variable against itself.

This is like a multiple regression but with lagged values of as predictors.
as predictors.
The two series in Figure 8.5 show series from an AR(1) model and an AR(2) model. Changing the parameters results in different time series patterns.
results in different time series patterns.
The variance of the error term will only change the scale of the series, not the patterns.
will only change the scale of the series, not the patterns. 8.1 Stationarity and differencing | Forecasting: Principles and Practice (2nd ed)
8.1 Stationarity and differencing | Forecasting: Principles and Practice (2nd ed)
For an AR(1) model:
when =0,
=0,  (or
(or  ) is equivalent to white noise;
) is equivalent to white noise;
when ϕ1=1 and c=0, (or
(or  ) is equivalent to a random walk;
) is equivalent to a random walk;

when ϕ1=1 and c≠0, is equivalent to a random walk with drift;
is equivalent to a random walk with drift;
 or
or 
The value of c is the average of the changes between consecutive observations
when ϕ1<0, (or
(or  ) tends to oscillate around the mean.
) tends to oscillate around the mean.
We normally restrict autoregressive models to stationary data, in which case some constraints on the values of the parameters are required.
For an AR(1) model :
:
−1< <1.
<1.
For an AR(2) model :
:
−1< <1, ϕ1+ϕ2<1, ϕ2−ϕ1<1.
<1, ϕ1+ϕ2<1, ϕ2−ϕ1<1.
When p≥3, the restrictions are much more complicated. - ARIMA(0,d,0):
First degree of differencing in the integrated component, also known as random walk, notated by I(d).
d is the degree of differencing, indicating the number of times the data have had past values subtracted表示数据减去过去值的次数.
For example, ARIMA(0,1,0) is I(1) and represented as follows:

Here, is the mean of the seasonal difference季节差分的平均值.
is the mean of the seasonal difference季节差分的平均值.
A seasonal difference is the difference between an observation and the previous observation from the same season. So

where m = the number of seasons. These are also called “lag-m differences”, as we subtract the observation after a lag of mm periods.
If seasonally differenced data appear to be white noise, then an appropriate model for the original data is

Forecasts from this model are equal to the last observation from the relevant season.
If OR
OR  denotes a seasonally differenced series, then the twice-differenced series is
denotes a seasonally differenced series, then the twice-differenced series is
When both seasonal and first differences
and first differences are applied, it makes no difference which is done first—the result will be the same. However, if the data have a strong seasonal pattern, we recommend that seasonal differencing be done first, because the resulting series will sometimes be stationary and there will be no need for a further first difference.因为结果序列有时会是平稳的,不需要进一步的一阶差分 If first differencing is done first, there will still be seasonality present如果先进行一阶差分,则仍然存在季节性.
are applied, it makes no difference which is done first—the result will be the same. However, if the data have a strong seasonal pattern, we recommend that seasonal differencing be done first, because the resulting series will sometimes be stationary and there will be no need for a further first difference.因为结果序列有时会是平稳的,不需要进一步的一阶差分 If first differencing is done first, there will still be seasonality present如果先进行一阶差分,则仍然存在季节性.
It is important that if differencing is used, the differences are interpretable.
First differences are the change between one observation and the next. S
easonal differences are the change between one year to the next.
Other lags are unlikely to make much interpretable sense and should be avoided. - ARIMA(0,0,q):
Moving average component, notated by MA(q).
The order q determines the number of terms to be included in the model:
Rather than using past values of the forecast variable in a regression, a moving average model uses past forecast errors in a regression-like model.
 OR
OR

 is white noise
is white noise
Of course, we do not observe the values of , so it is not really a regression in the usual sense.
, so it is not really a regression in the usual sense.
Notice that each value of can be thought of as a weighted moving average of the past few forecast errors. However, moving average models should not be confused with the moving average smoothing we discussed in Chapter 6. A moving average model is used for forecasting future values, while moving average smoothing is used for estimating the trend-cycle of past values.
can be thought of as a weighted moving average of the past few forecast errors. However, moving average models should not be confused with the moving average smoothing we discussed in Chapter 6. A moving average model is used for forecasting future values, while moving average smoothing is used for estimating the trend-cycle of past values. 8.1 Stationarity and differencing | Forecasting: Principles and Practice (2nd ed)
8.1 Stationarity and differencing | Forecasting: Principles and Practice (2nd ed)
Figure 8.6 shows some data from an MA(1) model and an MA(2) model.
Changing the parameters results in different time series patterns. As with autoregressive models,
results in different time series patterns. As with autoregressive models,
the variance of the error term will only change the scale of the series, not the patterns.
will only change the scale of the series, not the patterns.
It is possible to write any stationary AR(p) model as an MA( ) model
) model . For example, using repeated substitution, we can demonstrate this for an AR(1) model:
. For example, using repeated substitution, we can demonstrate this for an AR(1) model: and c=0 or the constant term c has been suppressed常数项已被抑制
and c=0 or the constant term c has been suppressed常数项已被抑制
Provided , the value of
, the value of  will get smaller as k gets larger. So eventually we obtain
will get smaller as k gets larger. So eventually we obtain an MA(
an MA(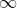 ) process.
) process.
The reverse result holds if we impose some constraints on the MA parameters. Then the MA model is called invertible. That is, we can write any invertible MA(q) process as an AR(∞) process. Invertible models are not simply introduced to enable us to convert from MA models to AR models. They also have some desirable mathematical properties.
For example, consider the MA(1) process, . In its AR(∞) representation, the most recent error can be written as a linear function of current and past observations:
. In its AR(∞) representation, the most recent error can be written as a linear function of current and past observations:
When , the weights increase as lags increase, so the more distant the observations the greater their influence on the current error.
, the weights increase as lags increase, so the more distant the observations the greater their influence on the current error.
When , the weights are constant in size, and the distant observations have the same influence as the recent observations. As neither of these situations make much sense, we require
, the weights are constant in size, and the distant observations have the same influence as the recent observations. As neither of these situations make much sense, we require  , so the most recent observations have higher weight than observations from the more distant past. Thus, the process is invertible when
, so the most recent observations have higher weight than observations from the more distant past. Thus, the process is invertible when  .
.
The invertibility constraints for other models are similar to the stationarity constraints
- For an MA(1) model:
 .
. - For an MA(2) model: −1<
 <1,
<1,  ,
,  .
.
More complicated conditions hold for .
.
- For an MA(1) model:
Finding model parameters by grid search
A grid search, also known as the hyperparameter optimization method, can be used to iteratively explore different combinations of parameters for fitting our ARIMA model. We can fit a seasonal ARIMA model with the SARIMAX() function of the statsmodels module in each iteration, returning an object of the MLEResults class. The MLEResults object holds an aic attribute for returning the AIC value. The model with the lowest AIC value gives us the best-fitting model that determines our parameters of p, d, and q. More information on SARIMAX can be found at statsmodels.tsa.statespace.sarimax.SARIMAX — statsmodels
We define the grid search procedure as the arima_grid_search() function, as follows:
import itertools
p = d = q = range(2)
list( itertools.product(p,d,q) ) 
statsmodels.tsa.statespace.sarimax.SARIMAX
Seasonal AutoRegressive Integrated Moving Average with eXogenous regressors model
You can see that we add P, D, and Q for the seasonal portion of the time series. They are the same terms as the non-seasonal components, by they involve backshifts of the seasonal period(m). In the formula above, m is the number of observations per year or the period. If we are analyzing quarterly data, m would equal 4.
Parameters:
-
endog array_like
The observed time-series process y
-
exog array_like,
optional
Array of exogenous/ ekˈsɑːdʒənəs /外生的,外因的,外成的 regressors, shaped nobs x k. -
order iterable or iterable
ofiterables,optionalThe (p,d,q) order of the model for the number of AR parameters, differences, and MA parameters. d must be an integer indicating the integration order of the process, while p and q may either be an integers indicating the AR and MA orders (so that all lags up to those orders are included) or else iterables giving specific AR and / or MA lags to include. Default is an AR(1) model: (1,0,0).
If we combine differencing with autoregression and a moving average model, we obtain a non-seasonal ARIMA model. ARIMA is an acronym for AutoRegressive Integrated Moving Average (in this context, “integration” is the reverse of differencing). The full model can be written as
MA(q)


AR(p) model :
Non-seasonal ARIMA models (8.1)
(8.1) -

where is the differenced series (it may have been differenced more than once可能不止一次差分). The “predictors” on the right hand side include both lagged values of
is the differenced series (it may have been differenced more than once可能不止一次差分). The “predictors” on the right hand side include both lagged values of  and lagged errors. We call this an ARIMA( p,d,q ) model, where
and lagged errors. We call this an ARIMA( p,d,q ) model, wherep= order of the autoregressive part; d= degree of first differencing involved; q= order of the moving average part. Once we start combining components in this way to form more complicated models, it is much easier to work with the backshift notation. For example, Equation (8.1) can be written in backshift notation(
 ,
,  ,
,  ) as
) as









-
seasonal_order iterable,
optionalThe (P,D,Q,s) order of the seasonal component of the model for the AR parameters, differences, MA parameters, and periodicity. D must be an integer indicating the integration order of the process, while P and Q may either be an integers indicating the AR and MA orders (so that all lags up to those orders are included) or else iterables giving specific AR and / or MA lags to include. s is an integer giving the periodicity (number of periods in season), often it is 4 for quarterly data or 12 for monthly data. Default is no seasonal effect.
VVVVVVVVVV
In the case of a SARIMA model with only a seasonal moving average process of order 1(Q=1) and period of 12, denoted as:
Moving average component, notated by MA(q).
The order q determines the number of terms to be included in the model:
Rather than using past values of the forecast variable in a regression, a moving average model uses past forecast errors in a regression-like model.
 or
or

^^^^^^^^^^^^^^Similarly, for a model with only a seasonal autoregressive process of order 1 and period of 12:

############
ARIMA(p,0,0):
First-order autoregressive model, notated by AR(p).
p is the lag order, indicating the number of lagged observations in the model.
AR(1) :
 or
or 
For example, ARIMA(2,0,0) is AR(2) and represented as follows:

Here, and
and  are parameters for the model. and
are parameters for the model. and  is white noise
is white noise
In a multiple regression model, we forecast the variable of interest using a linear combination of predictors. In an autoregression model, we forecast the variable of interest using a linear combination of past values of the variable
of interest using a linear combination of past values of the variable . The term autoregression indicates that it is a regression of the variable against itself.
. The term autoregression indicates that it is a regression of the variable against itself.

This is like a multiple regression but with lagged values of as predictors.
as predictors.############
-
trend str{‘n’,’c’,’t’,’ct’}
oriterable,optionalParameter controlling the deterministic trend polynomial A(t). Can be specified as a string where ‘c’ indicates a constant (i.e. a degree zero component of the trend polynomial), ‘t’ indicates a linear trend with time, and ‘ct’ is both. Can also be specified as an iterable defining the non-zero polynomial exponents to include, in increasing order. For example, [1,1,0,1] denotes
 . Default is to not include a trend component.
. Default is to not include a trend component.
Notes
The SARIMA model is specified ![]() .
. ![]()
- AR(P) :

 是非季节性自回归滞后多项式 :捕获非季节性自回归因素。
是非季节性自回归滞后多项式 :捕获非季节性自回归因素。
 是季节性自回归滞后多项式:捕获季节性回归因素。
是季节性自回归滞后多项式:捕获季节性回归因素。
 是时序数据 d 阶差分;季节性D阶差分:提供时序平稳化功能。
是时序数据 d 阶差分;季节性D阶差分:提供时序平稳化功能。- A(t) 为趋势多项式:包括截距项
- MA(q) :

 是非季节性移动平均滞后多项式
是非季节性移动平均滞后多项式
 是季节性移动平均滞后多项式
是季节性移动平均滞后多项式
For example, an ARIMA![]() model (without a constant c = A(t)=0 ) is for quarterly data (s=4), and can be written as
model (without a constant c = A(t)=0 ) is for quarterly data (s=4), and can be written as
![]()
![]()
For example, an ARIMA![]() model (no differencing, no AR terms, no constant c = A(t)=0)
model (no differencing, no AR terms, no constant c = A(t)=0)![]()
![]()
For example, an ARIMA![]() model (no differencing, no MA terms, no constant c = A(t)=0)
model (no differencing, no MA terms, no constant c = A(t)=0)![]()
For example, an ARIMA![]() model( A(t)=c, 意味着趋势多项式就是一个截距项)
model( A(t)=c, 意味着趋势多项式就是一个截距项)![]()

![]()
![]()
![]()
![]()
d=1,D=1,s=12 意味着 ![]() 是经过一阶差分和季节性(12个月)差分所得到的
是经过一阶差分和季节性(12个月)差分所得到的
In terms of a univariate structural model单变量结构模型, this can be represented as![]()
where ![]() is only applicable in the case of measurement error (although it is also used in the case of a pure regression model, i.e. if p=q=0).
is only applicable in the case of measurement error (although it is also used in the case of a pure regression model, i.e. if p=q=0).
In terms of this model, regression with SARIMA errors can be represented easily as ![]()
this model is the one used when exogenous regressors are provided.
import itertools
import warnings
from statsmodels.tsa.statespace.sarimax import SARIMAX
warnings.filterwarnings("ignore")
def arima_grid_search( dataframe, periods ):
p = d = q = range(2)
param_combinations = list( itertools.product(p,d,q) )
lowest_aic, pdq, pdqs = None, None, None
total_iterations = 0
for order in param_combinations: # order ==> non_seasonal
for (p,q,d) in param_combinations:
seasonal_order = (p,q,d,periods)
total_iterations += 1
try:
model = SARIMAX( df_settle, order= order,
seasonal_order=seasonal_order,
enforce_stationarity = False,
enforce_invertability = False,
disp = False
)
model_result = model.fit( maxiter=200, disp=False )
if not lowest_aic or model_result.aic < lowest_aic:
lowest_aic = model_result.aic
pdq, pdqs = order, seasonal_order
except Exception as ex:
continue
return lowest_aic, pdq, pdqsOur variable, df_settle , holds the monthly prices of the futures data that we downloaded in the previous section. In the SARIMAX (seasonal autoregressive integrated moving average with exogenous/ ekˈsɑːdʒənəs /外生的 regressors model) function, we provided the seasonal_order parameter, which is the ARIMA(p,d,q,s) seasonal component, where s is the number of periods in a season of the dataset. Since we are using monthly data, we use 12 periods to define a seasonal pattern. The enforce_stationarity=False parameter doesn't transform the AR parameters to enforce stationarity in the AR component of the model. The enforce_invertibility=False parameter doesn't transform MA parameters to enforce invertibility in the MA component of the model. The disp=False parameter suppresses output information when fitting our models.
With the grid function defined, we can now call this with our monthly data and print out the model parameters with the lowest AIC value:
lowest_aic, order, seasonal_order = arima_grid_search( df_settle, 12 )
print( 'ARIMA{}X{}'.format(order, seasonal_order) )
print( 'Lowest AIC: %.3f' % lowest_aic )![]()
An ARIMA(0, 1, 1, 12) seasonal component model would give us the lowest AIC value at 2536.884. We shall use these parameters to fit our SARIMAX model in the next section.
from statsmodels.tsa.arima.model import ARIMA
model = ARIMA( model_results.resid, order=order, seasonal_order=seasonal_order )
fitted_results = model.fit()
model_results.resid.plot()
fitted_results.fittedvalues.plot( color='red' )
plt.setp( plt.gca().get_xticklabels(), rotation=30, horizontalalignment='right' )
plt.show()AIC and BIC for SARIMAX
Akaike’s Information Criterion (AIC), which was useful in selecting predictors for regression, is also useful for determining the order of an ARIMA model. It can be written as
![]() ==>
==> ![]()
where L is the likelihood of the data
(![]() is the maximized value of the likelihood function of the model M. i.e.
is the maximized value of the likelihood function of the model M. i.e. ![]() , where
, where ![]() are the parameter values that maximize the likelihood function,
are the parameter values that maximize the likelihood function, ![]() = the observed data;
= the observed data;
), k=1 if c≠0 and k=0 if c=0. Note that the last term in parentheses is the number of parameters in the model (including ![]() , the variance of the residuals).
, the variance of the residuals).
For ARIMA models, the corrected AIC can be written as 
and the Bayesian Information Criterion(![]() ) can be written as
) can be written as ![]()
It is important to note that these information criteria tend not to be good guides to selecting the appropriate order of differencing (d) of a model, but only for selecting the values of p and q. This is because the differencing changes the data on which the likelihood is computed, making the AIC values between models with different orders of differencing not comparable. So we need to use some other approach to choose d, and then we can use the AICc to select p and q.
Fitting the SARIMAX model
Having obtained the optimal model parameters, inspect the model properties using the summary() method on the fitted results to view detailed statistical information:
model = SARIMAX(
df_settle,
order=order,
seasonal_order = seasonal_order,
enforce_stationarity = False,
endorce_invertibility = False,
disp=False
)
model_results = model.fit( maxiter=200, disp=False)
print( model_results.summary() )![]()
It is important to run model diagnostics to investigate that model assumptions haven't been
violated:
The top-right plot shows the kernel density estimate (KDE) of the standardized residuals, which suggests the errors are Gaussian with a mean close to zero. Let's see a more accurate statistic of the residuals:
model_results.resid.describe() ![]() From the description of the residuals, the non-zero mean suggests that the prediction may be biased positively.
From the description of the residuals, the non-zero mean suggests that the prediction may be biased positively.
Predicting and forecasting the SARIMAX model
The model_results variable is a SARIMAXResults object of the statsmodel module, representing the output of the SARIMAX model. It contains a get_prediction() method for performing in-sample prediction and out-of-sample forecasting. It also contains a conf_int() method, which returns the confidence intervals of the predictions, both lower- and upper-bounded, of the fitted parameters, which is at a 95% confidence interval by default. Let's apply these methods:
n = len( df_settle.index ) # number of months
# start = n-12*5 : in-sample prediction of the most recent five years' prices
# n+5 : out-of-sample forecast of the next five months.
prediction = model_results.get_prediction( start = n-12*5,
end = n+5
)
prediction_ci = prediction.conf_int()The start parameter in the get_prediction() method indicates we are performing an in-sample prediction of the most recent five years' prices. At the same time, with the end parameter, we are performing an out-of-sample forecast of the next five months
By inspecting the top five forecasted confidence interval values, we get the following:
print( prediction_ci.head(5) )Let's plot the predicted and forecasted prices against our original dataset, from the year 2008 onwards:
df_settle.indexplt.figure(figsize=(12, 6))
ax = df_settle['2008':].plot(label='actual')
prediction_ci.plot( ax=ax, style=['--', '--'],
label='predicted/forecasted')
ci_index = prediction_ci.index
lower_ci = prediction_ci.iloc[:, 0]
upper_ci = prediction_ci.iloc[:, 1]
ax.fill_between( ci_index,
lower_ci, upper_ci,
color='r', alpha=.1
)
ax.set_xlabel('Time (years)')
ax.set_ylabel('Prices')
plt.legend()
plt.show() 
The solid line plot shows the observed values, while the dotted lines plot the five-year rolling predictions trailing closely and bounded by the confidence intervals in the shaded area. Observe that as the next five-month forecast goes into the future, and the confidence interval widens to reflect the loss of certainty in the outlook.
Summary
In this chapter, we were introduced to PCA as a dimension reduction technique in portfolio modeling. By breaking down the movement of asset prices of a portfolio into its principal components, or common factors, the most useful factors can be kept, and portfolio analysis can be greatly simplified without compromising on computational time and space complexity. In applying PCA to the Dow and its thirty components using the KernelPCA function of the sklearn.decomposition module, we obtained eigenvectors and eigenvalues, which we used to reconstruct the Dow with five components.
In the statistical analysis of time series data, the data is considered as either stationary or non-stationary. Stationary time series data is data whose statistical properties are constant
over time. Non-stationary time series data has its statistical properties change over time, most likely due to trends, seasonality, presence of a unit root, or a combination of all three. Modeling from non-stationary data may produce spurious regression. In order to receive consistent and reliable results, non-stationary data needs to be transformed into stationary data.
We used statistical tests such as the ADF(Augmented Dickey-Fuller Test) to check whether stationary expectations are met or violated. The adfuller method of the statsmodels.tsa.stattools module provides the test statistic, p-value, and critical values, from which we can fail to reject the null hypothesis that the data has a unit root and is non-stationary.
We transformed non-stationary data into stationary data by detrending, differencing, and seasonal decomposition. By using ARIMA, we fitted models using the SARIMAX function of the statsmodels.tsa.statespace.sarimax module to find suitable model parameters that give the lowest AIC value through an iterative grid search procedure. The fitted results are used for prediction and forecasting.
In the next chapter, we will perform interactive financial analytics with the VIX(Volatility Index, measures the short-term volatility implied by S&P 500 stock index options with an average expiration of 30 days).













































