limma差异分析谁比谁不重要
文章目录
-
- 准备数据
- 没有比较矩阵
- 用比较矩阵
- 总结
新手在刚接触
limma包做差异分析的时候,会碰到很多教程,有的教程写的是 正常组比疾病组,有的是 疾病组比正常组,他们都是对的,只有你凌乱了。
其实无所谓,你要记住:如果normal比tumor是高表达,logfc是正的,那么tumor比normal就是低表达,logfc是负的。用的时候要搞清楚到底是谁比谁!
下面用一个例子说下limma的逻辑。
准备数据
这个数据一共17个样本,前7个是正常(normal)组,后10个是溃疡性结肠炎(uc)组。
rm(list = ls())
load(file = '../000files/step1-output.Rdata')
# 表达矩阵
exprSet[1:4,1:4]
## GSM901319 GSM901320 GSM901321 GSM901322
## IGLC1 13.98084 14.49569 14.01361 14.14011
## RPL41 14.44795 14.35745 14.46906 14.31751
## ND4 14.49569 14.23311 14.54316 14.46906
## EEF1A1 14.52687 14.39076 14.34238 14.27464
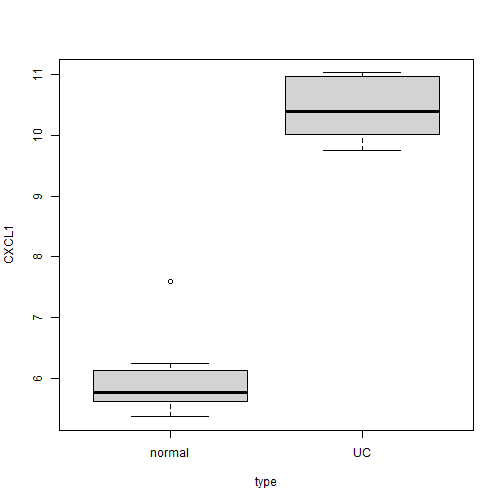
为了方便大家理解,现在我们先挑一个CXCL1这个基因,根据背景知识,这个基因在uc组绝壁是高表达!我们可以画个箱线图看一下:
tmp <- as.data.frame(t(exprSet["CXCL1",]))
tmp$type <- group_list
boxplot(CXCL1 ~ type, data = tmp)
箱线图和我们的背景知识也是一致的。如果是UC比normal,那肯定是高表达,logFC应该是正的;如果是normal比UC,那肯定是低表达,logFC应该是负的。
用limma做差异分析非常灵活,你可以用比较矩阵,也可以不用比较矩阵。
没有比较矩阵
多于两个分组的不要用这种方法。
定义下分组,这个分组和我们表达矩阵列名是对应的,前7个normal,后10个uc。
group_list <- c(rep('normal',7),rep('uc',10))
group_list
## [1] "normal" "normal" "normal" "normal" "normal" "normal" "normal" "uc"
## [9] "uc" "uc" "uc" "uc" "uc" "uc" "uc" "uc"
## [17] "uc"
注意!这时候
limma包默认是排序靠后的 vs 排序靠前的!。
比如,现在我们的group_list还是字符串,这时候的默认顺序是 normal在前,uc在后,这时候你的设计矩阵design是这样的:
library(limma)
# 用不用factor()都不影响,必定是 靠后的 vs 靠前的
design <- model.matrix(~ group_list) # 这里 没有 0 哦!!
design
## (Intercept) group_listuc
## 1 1 0
## 2 1 0
## 3 1 0
## 4 1 0
## 5 1 0
## 6 1 0
## 7 1 0
## 8 1 1
## 9 1 1
## 10 1 1
## 11 1 1
## 12 1 1
## 13 1 1
## 14 1 1
## 15 1 1
## 16 1 1
## 17 1 1
## attr(,"assign")
## [1] 0 1
## attr(,"contrasts")
## attr(,"contrasts")$group_list
## [1] "contr.treatment"
注意观察group_listuc那一列,这时候uc是1,normal是0,所以这种情况下继续往下做差异分析肯定是uc vs normal,CXCL1的logFC绝壁是正的!
差异分析:
fit <- lmFit(exprSet, design)
fit <- eBayes(fit)
allDiff <- topTable(fit, number = Inf)
## Removing intercept from test coefficients
head(allDiff,12)
## logFC AveExpr t P.Value adj.P.Val B
## HMGCS2 -6.018195 8.483645 -23.68806 6.183283e-15 1.247415e-10 23.77686
## CHI3L1 6.532374 8.423055 22.34265 1.699896e-14 1.714685e-10 22.90827
## SLC26A2 -5.096618 10.384007 -21.30421 3.860702e-14 2.596194e-10 22.19093
## CLDN8 -6.408782 7.228457 -18.83007 3.197251e-13 1.612534e-09 20.29435
## S100A8 6.039988 8.849455 17.57688 1.029574e-12 4.154125e-09 19.21875
## CDC25B 1.703420 8.246338 16.73922 2.347422e-12 7.892815e-09 18.45069
## DPP10 -2.020786 3.617608 -16.36731 3.424769e-12 8.132674e-09 18.09612
## PDPN 2.821963 6.311157 16.31720 3.605676e-12 8.132674e-09 18.04767
## DPP10-AS1 -1.665377 4.529724 -16.21688 3.998717e-12 8.132674e-09 17.95022
## CFB 2.878172 8.304561 16.19818 4.076831e-12 8.132674e-09 17.93199
## CXCL1 4.418606 8.627389 16.11715 4.434391e-12 8.132674e-09 17.85270
## AQP8 -6.916403 7.588945 -15.82926 5.996104e-12 9.395533e-09 17.56754
可以看到CXCL1的logFC是正的,是高表达的,没有任何问题。下面再画一个火山图看一看。
library(ggplot2)
library(ggrepel)
allDiff$type <- ifelse(allDiff$logFC > 2 & allDiff$adj.P.Val < 0.01, "up",
ifelse(allDiff$logFC < -2 & allDiff$adj.P.Val < 0.01, "down", "not-sig")
)
allDiff$gene <- rownames(allDiff)
p1 <- ggplot(allDiff, aes(logFC, -log10(adj.P.Val)))+
geom_point(aes(color=type))+
scale_color_manual(values = c("blue","black","red"))+
geom_hline(yintercept = -log10(0.01),linetype=2)+
geom_vline(xintercept = c(-2,2), linetype=2)+
geom_text_repel(data = subset(allDiff, abs(logFC) > 4),
aes(label=gene),col="black",alpha = 0.8)+
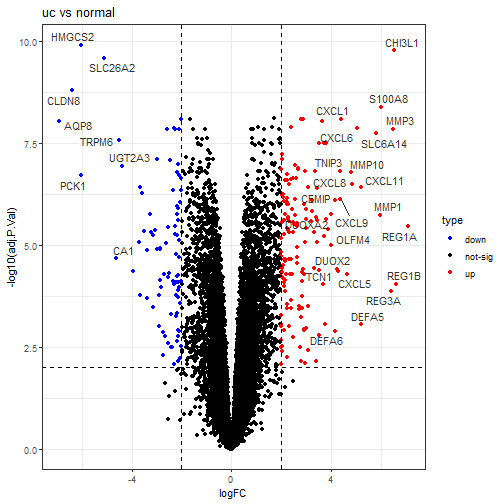
ggtitle("uc vs normal")+
theme_bw()
p1
在这个火山图里,CXCL1是高表达,现在是 uc 比 normal,没有任何问题,和我们的背景知识以及箱线图都一致!
下面我们把分组稍微修改一下,把normal改成znormal,这时在R语言里面默认顺序就变成uc在前,znormal在后。
group_list <- c(rep('znormal',7),rep('uc',10))
group_list
## [1] "znormal" "znormal" "znormal" "znormal" "znormal" "znormal" "znormal"
## [8] "uc" "uc" "uc" "uc" "uc" "uc" "uc"
## [15] "uc" "uc" "uc"
这时候的design也会跟着变化:
design <- model.matrix(~ group_list)
design
## (Intercept) group_listznormal
## 1 1 1
## 2 1 1
## 3 1 1
## 4 1 1
## 5 1 1
## 6 1 1
## 7 1 1
## 8 1 0
## 9 1 0
## 10 1 0
## 11 1 0
## 12 1 0
## 13 1 0
## 14 1 0
## 15 1 0
## 16 1 0
## 17 1 0
## attr(,"assign")
## [1] 0 1
## attr(,"contrasts")
## attr(,"contrasts")$group_list
## [1] "contr.treatment"
看清楚了吗?这时候后面一列是group_listznormal了,而且znormal是1,uc是0!
此时你往下做差异分析,刚好和上面完全反过来!CXCL1的logFC会变成负的,因为此时是znormal vs uc!
fit <- lmFit(exprSet,design)
fit <- eBayes(fit)
allDiff <- topTable(fit, number = Inf)
## Removing intercept from test coefficients
head(allDiff, 12)
## logFC AveExpr t P.Value adj.P.Val B
## HMGCS2 6.018195 8.483645 23.68806 6.183283e-15 1.247415e-10 23.77686
## CHI3L1 -6.532374 8.423055 -22.34265 1.699896e-14 1.714685e-10 22.90827
## SLC26A2 5.096618 10.384007 21.30421 3.860702e-14 2.596194e-10 22.19093
## CLDN8 6.408782 7.228457 18.83007 3.197251e-13 1.612534e-09 20.29435
## S100A8 -6.039988 8.849455 -17.57688 1.029574e-12 4.154125e-09 19.21875
## CDC25B -1.703420 8.246338 -16.73922 2.347422e-12 7.892815e-09 18.45069
## DPP10 2.020786 3.617608 16.36731 3.424769e-12 8.132674e-09 18.09612
## PDPN -2.821963 6.311157 -16.31720 3.605676e-12 8.132674e-09 18.04767
## DPP10-AS1 1.665377 4.529724 16.21688 3.998717e-12 8.132674e-09 17.95022
## CFB -2.878172 8.304561 -16.19818 4.076831e-12 8.132674e-09 17.93199
## CXCL1 -4.418606 8.627389 -16.11715 4.434391e-12 8.132674e-09 17.85270
## AQP8 6.916403 7.588945 15.82926 5.996104e-12 9.395533e-09 17.56754
CXCL1的logFC是负的了,没有问题吧?画火山图也是完全相反的。
allDiff$type <- ifelse(allDiff$logFC > 2 & allDiff$adj.P.Val < 0.01, "up",
ifelse(allDiff$logFC < -2 & allDiff$adj.P.Val < 0.01, "down", "not-sig")
)
allDiff$gene <- rownames(allDiff)
p2 <- ggplot(allDiff, aes(logFC, -log10(adj.P.Val)))+
geom_point(aes(color=type))+
scale_color_manual(values = c("blue","black","red"))+
geom_hline(yintercept = -log10(0.01),linetype=2)+
geom_vline(xintercept = c(-2,2), linetype=2)+
geom_text_repel(data = subset(allDiff, abs(logFC) > 4),
aes(label=gene),col="black",alpha = 0.8)+
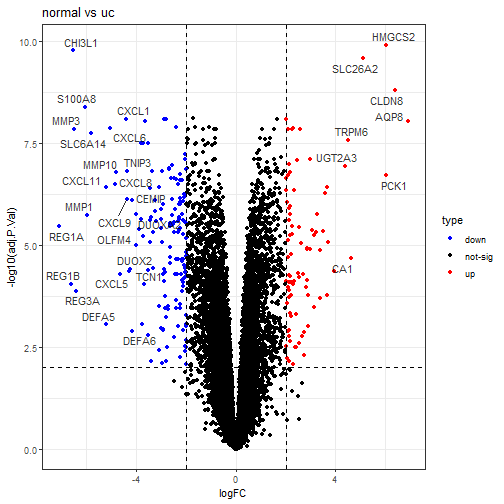
ggtitle("normal vs uc")+
theme_bw()
p2
可以看到CXCL1是低表达的,有问题吗?完全没问题,因为此时是znormal vs uc,是正常组比uc组,肯定是低的了。
完全相反,但是完全没有问题。
如果你喜欢在设计矩阵时喜欢用
factor()函数,比如design <- model.matrix(~ factor(group_list)),也是一样的道理,顺序靠后的 vs 顺序靠前的。
group_list <- c(rep('znormal',7),rep('uc',10))
# 定义因子顺序,让zormal在前,uc在后
design <- model.matrix(~ factor(group_list, levels = c("znormal","uc")))
design
## (Intercept) factor(group_list, levels = c("znormal", "uc"))uc
## 1 1 0
## 2 1 0
## 3 1 0
## 4 1 0
## 5 1 0
## 6 1 0
## 7 1 0
## 8 1 1
## 9 1 1
## 10 1 1
## 11 1 1
## 12 1 1
## 13 1 1
## 14 1 1
## 15 1 1
## 16 1 1
## 17 1 1
## attr(,"assign")
## [1] 0 1
## attr(,"contrasts")
## attr(,"contrasts")$`factor(group_list, levels = c("znormal", "uc"))`
## [1] "contr.treatment"
看到了吗,这个矩阵还是靠后(这里是uc)的是1,所以往下做差异分析还是uc vs znormal,结果CXCL1的logFC绝壁是正的!
用比较矩阵
这时候决定到底是谁和谁比的,是你的比较矩阵,其他的顺序都无所谓了!
group_list <- c(rep('normal',7),rep('uc',10))
group_list
## [1] "normal" "normal" "normal" "normal" "normal" "normal" "normal" "uc"
## [9] "uc" "uc" "uc" "uc" "uc" "uc" "uc" "uc"
## [17] "uc"
# 用不用factor()都无所谓
design <- model.matrix(~ 0 + group_list)
design
## group_listnormal group_listuc
## 1 1 0
## 2 1 0
## 3 1 0
## 4 1 0
## 5 1 0
## 6 1 0
## 7 1 0
## 8 0 1
## 9 0 1
## 10 0 1
## 11 0 1
## 12 0 1
## 13 0 1
## 14 0 1
## 15 0 1
## 16 0 1
## 17 0 1
## attr(,"assign")
## [1] 1 1
## attr(,"contrasts")
## attr(,"contrasts")$group_list
## [1] "contr.treatment"
改个名字,更好看一点:
colnames(design) <- c("normal","uc")
rownames(design) <- colnames(exprSet)
design
## normal uc
## GSM901319 1 0
## GSM901320 1 0
## GSM901321 1 0
## GSM901322 1 0
## GSM901323 1 0
## GSM901324 1 0
## GSM901325 1 0
## GSM901339 0 1
## GSM901341 0 1
## GSM901342 0 1
## GSM901344 0 1
## GSM901346 0 1
## GSM901347 0 1
## GSM901348 0 1
## GSM901349 0 1
## GSM901350 0 1
## GSM901351 0 1
## attr(,"assign")
## [1] 1 1
## attr(,"contrasts")
## attr(,"contrasts")$group_list
## [1] "contr.treatment"
下面我们定义一个比较矩阵,明确到底要谁和谁比!
# 明确 uc vs normal
contrast.matrix <- makeContrasts(uc - normal, levels = design)
contrast.matrix
## Contrasts
## Levels uc - normal
## normal -1
## uc 1
进行差异分析:
fit <- lmFit(exprSet, design)
fit <- contrasts.fit(fit, contrast.matrix)
fit <- eBayes(fit)
allDiff <- topTable(fit, number = Inf)
head(allDiff, 12)
## logFC AveExpr t P.Value adj.P.Val B
## HMGCS2 -6.018195 8.483645 -23.68806 6.183283e-15 1.247415e-10 23.77686
## CHI3L1 6.532374 8.423055 22.34265 1.699896e-14 1.714685e-10 22.90827
## SLC26A2 -5.096618 10.384007 -21.30421 3.860702e-14 2.596194e-10 22.19093
## CLDN8 -6.408782 7.228457 -18.83007 3.197251e-13 1.612534e-09 20.29435
## S100A8 6.039988 8.849455 17.57688 1.029574e-12 4.154125e-09 19.21875
## CDC25B 1.703420 8.246338 16.73922 2.347422e-12 7.892815e-09 18.45069
## DPP10 -2.020786 3.617608 -16.36731 3.424769e-12 8.132674e-09 18.09612
## PDPN 2.821963 6.311157 16.31720 3.605676e-12 8.132674e-09 18.04767
## DPP10-AS1 -1.665377 4.529724 -16.21688 3.998717e-12 8.132674e-09 17.95022
## CFB 2.878172 8.304561 16.19818 4.076831e-12 8.132674e-09 17.93199
## CXCL1 4.418606 8.627389 16.11715 4.434391e-12 8.132674e-09 17.85270
## AQP8 -6.916403 7.588945 -15.82926 5.996104e-12 9.395533e-09 17.56754
结果和预想的一样。火山图就不画了。
如果是明确 normal vs uc,那结果肯定是相反的:
contrast.matrix <- makeContrasts(normal - uc, levels = design)
fit <- lmFit(exprSet, design)
fit <- contrasts.fit(fit, contrast.matrix)
fit <- eBayes(fit)
allDiff <- topTable(fit, number = Inf)
head(allDiff, 12)
## logFC AveExpr t P.Value adj.P.Val B
## HMGCS2 6.018195 8.483645 23.68806 6.183283e-15 1.247415e-10 23.77686
## CHI3L1 -6.532374 8.423055 -22.34265 1.699896e-14 1.714685e-10 22.90827
## SLC26A2 5.096618 10.384007 21.30421 3.860702e-14 2.596194e-10 22.19093
## CLDN8 6.408782 7.228457 18.83007 3.197251e-13 1.612534e-09 20.29435
## S100A8 -6.039988 8.849455 -17.57688 1.029574e-12 4.154125e-09 19.21875
## CDC25B -1.703420 8.246338 -16.73922 2.347422e-12 7.892815e-09 18.45069
## DPP10 2.020786 3.617608 16.36731 3.424769e-12 8.132674e-09 18.09612
## PDPN -2.821963 6.311157 -16.31720 3.605676e-12 8.132674e-09 18.04767
## DPP10-AS1 1.665377 4.529724 16.21688 3.998717e-12 8.132674e-09 17.95022
## CFB -2.878172 8.304561 -16.19818 4.076831e-12 8.132674e-09 17.93199
## CXCL1 -4.418606 8.627389 -16.11715 4.434391e-12 8.132674e-09 17.85270
## AQP8 6.916403 7.588945 15.82926 5.996104e-12 9.395533e-09 17.56754
可以看到CXCL1的logFC变成负的了。
总结
- 如果不用比较矩阵,就是默认 顺序靠后的 vs 顺序靠前的;
- 如果用比较矩阵,那就是按照你定义的进行比较,你让它怎么比它就怎么比;
- 推荐大家用比较矩阵,明确谁和谁比,不容易出错!