【打卡】糖尿病遗传风险检测挑战赛
赛题背景
截至2022年,中国糖尿病患者近1.3亿。中国糖尿病患病原因受生活方式、老龄化、城市化、家族遗传等多种因素影响。同时,糖尿病患者趋向年轻化。
糖尿病可导致心血管、肾脏、脑血管并发症的发生。因此,准确诊断出患有糖尿病个体具有非常重要的临床意义。糖尿病早期遗传风险预测将有助于预防糖尿病的发生。
根据《中国2型糖尿病防治指南(2017年版)》,糖尿病的诊断标准是具有典型糖尿病症状(烦渴多饮、多尿、多食、不明原因的体重下降)且随机静脉血浆葡萄糖≥11.1mmol/L或空腹静脉血浆葡萄糖≥7.0mmol/L或口服葡萄糖耐量试验(OGTT)负荷后2h血浆葡萄糖≥11.1mmol/L。
在这次比赛中,您需要通过训练数据集构建糖尿病遗传风险预测模型,然后预测出测试数据集中个体是否患有糖尿病,和我们一起帮助糖尿病患者解决这“甜蜜的烦恼”。
Task1:比赛报名
data_train = pd.read_csv('./比赛训练集.csv',encoding='gbk')
data_test = pd.read_csv('./比赛测试集.csv',encoding='gbk')
print('训练集',data_train.shape)
print('测试集',data_test.shape)
训练集 (5070, 10)
测试集 (1000, 9)
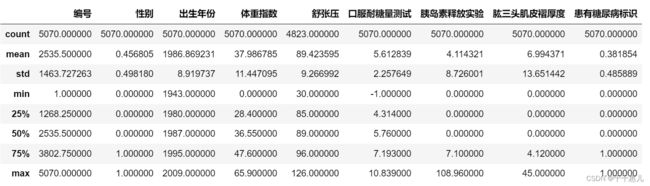
data_train.describe()
Task2:比赛数据分析
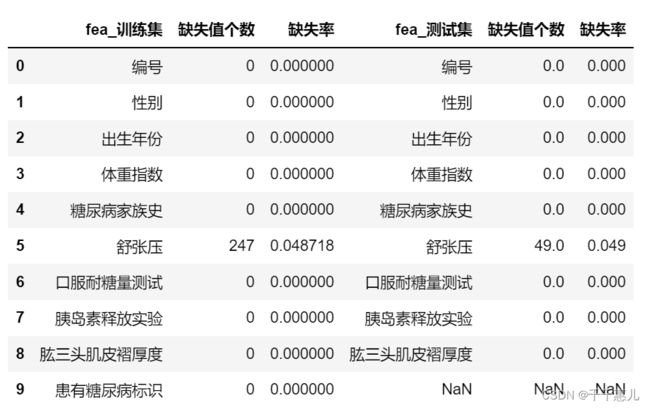
2.1缺失值
训练集和测试集都只有【舒张压】有缺失,训练集和测试集缺失比例差别不大
fea_lst = []
num_lst = []
num_rate_lst = []
for i in list(data_train.columns):
num = data_train[i].isnull().sum()
num_rate = num/len(data_train)
fea_lst.append(i)
num_lst.append(num)
num_rate_lst.append(num_rate)
df_null_ana_train = pd.DataFrame({'fea_训练集':fea_lst,'缺失值个数':num_lst,'缺失率':num_rate_lst})
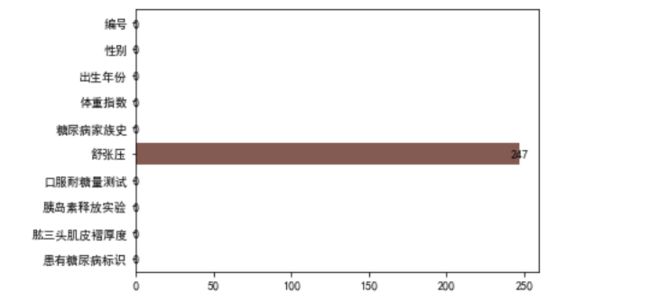
#各特征缺失数量统计
null_count = data_train.isnull().sum().values
# 缺失值情况
plt.figure()
sns.barplot(x = null_count, y = data_train.columns)
for x, y in enumerate(null_count):
plt.text(y, x, "%s" %y, horizontalalignment='center', verticalalignment='center')
plt.show()
2.2 数据类型
data_train.dtypes
编号 int64
性别 int64
出生年份 int64
体重指数 float64
糖尿病家族史 object
舒张压 float64
口服耐糖量测试 float64
胰岛素释放实验 float64
肱三头肌皮褶厚度 float64
患有糖尿病标识 int64
连续型变量:出生年份、体重指数、舒张压、口服耐糖量测试、胰岛素释放实验、肱三头肌皮褶厚度
类别型变量:【性别】【糖尿病家族史】,分布如下:

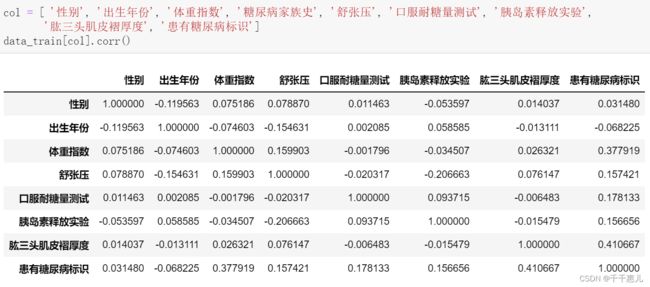
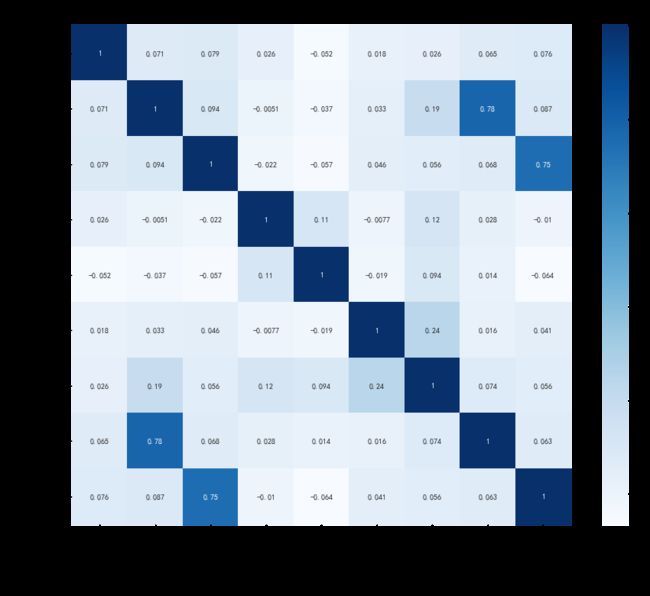
2.3 相关性分析
plt.rcParams['font.sans-serif'] = ['SimHei'] # 正常显示中文
plt.rcParams['axes.unicode_minus'] = False
plt.subplots(figsize=(15, 12)) # 设置画面大小
plt.title('Correlation between features', fontsize = 30)
sns.heatmap(data[column].corr(), annot=True, vmax=1, square=True, cmap="Blues")
plt.savefig('./相关系数热力图.png')
plt.show()
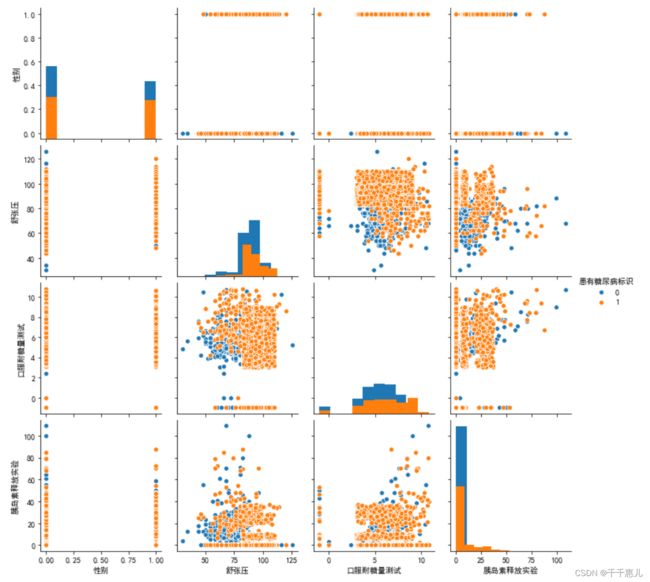
使用其他可视化方法将字段 与 标签的分布差异进行可视化
sns.pairplot(data_train,diag_kind='hist',hue='患有糖尿病标识')
plt.show()
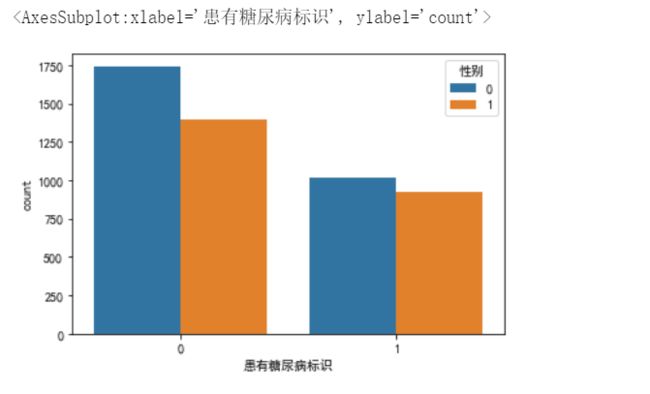
data_train['性别'].value_counts().plot(kind='bar') # 柱形图,plot是pandas封装的函数,底层是matplotlib
# data_train['性别'].value_counts().plot(kind='barh') # 条形图
sns.countplot(x='患有糖尿病标识',hue='性别',data=data_train)
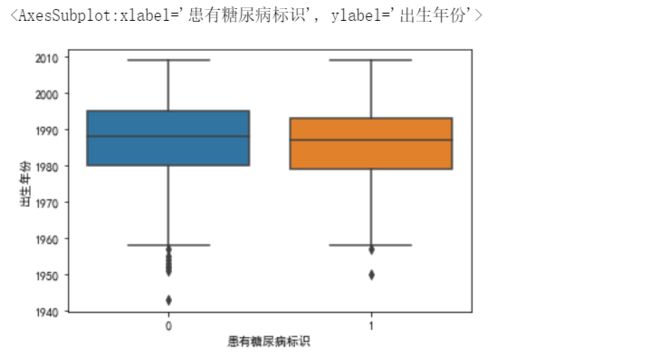
sns.boxplot(y='出生年份',x='患有糖尿病标识',data=data_train)
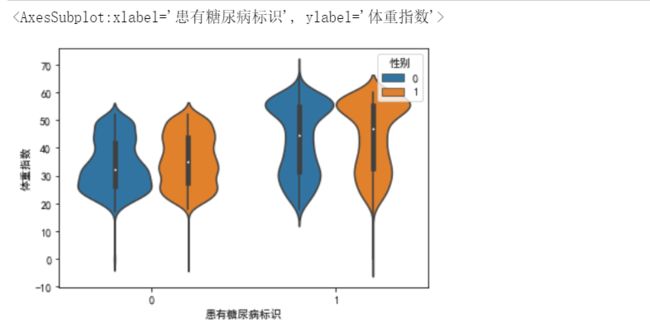
sns.boxplot(y='体重指数',x='患有糖尿病标识',hue='性别',data=data_train)
从下图可以看出,患有糖尿病的群体,体重指数整体是偏高的

小提琴图
sns.violinplot(y='体重指数',x='患有糖尿病标识',hue='性别',data=data_train)
Task3:逻辑回归
from sklearn.metrics import confusion_matrix
from sklearn.linear_model import LogisticRegression as LR
lr = LR(C=1e2,random_state=2020)
train_x = df_train[col].drop('患有糖尿病标识',axis=1).values
train_y = df_train['患有糖尿病标识'].values
lr.fit(train_x,train_y)
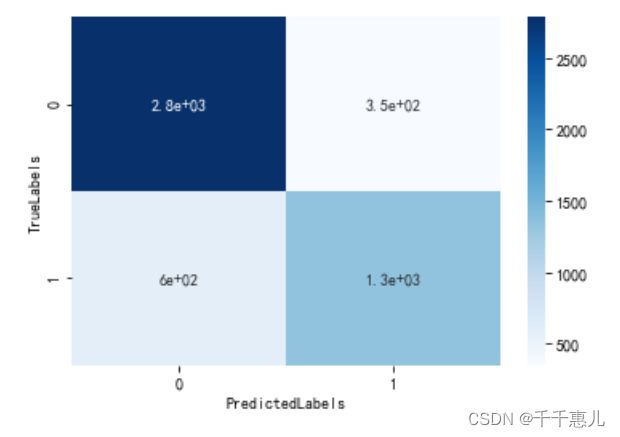
result = confusion_matrix(train_y, lr.predict(train_x))
sns.heatmap(result, annot=True, cmap='Blues')
plt.xlabel('PredictedLabels')
plt.ylabel('TrueLabels')
plt.show()
Task4:特征工程
思路
1、缺失值填充:口服耐糖量测试
2、特征衍生:统计每个性别对应的【体重指数】、【舒张压】平均值;计算每个患者与每个性别平均值的差异;
3、特征编码:糖尿病家族史
4、业务含义:年龄、体重指数、舒张压
df_ana = pd.DataFrame(data['性别'].value_counts()).reset_index()
df_ana.columns = ['性别','客户量']
for i in ['体重指数','舒张压']:
temp = data.groupby(['性别'],as_index=False)[i].agg({'mean','std'}).reset_index()
temp.columns = ['性别','{}_mean'.format(i),'{}_std'.format(i)]
df_ana = pd.merge(df_ana,temp,how='left',on='性别')
df_ana
data['舒张压'] = data['舒张压'].fillna(0)
data['体重指数_round'] = data['体重指数'] // 10
data['年龄'] = 2022 - data['出生年份']
data['口服耐糖量测试'] = data['口服耐糖量测试'].replace(-1, np.nan)
dict_糖尿病家族史 = {
'无记录': 0,
'叔叔或姑姑有一方患有糖尿病': 1,
'叔叔或者姑姑有一方患有糖尿病': 1,
'父母有一方患有糖尿病': 2
}
data['糖尿病家族史'] = data['糖尿病家族史'].map(dict_糖尿病家族史)
def BMI(a):
"""
人体的成人体重指数正常值是在18.5-24之间
低于18.5是体重指数过轻
在24-27之间是体重超重
27以上考虑是肥胖
高于32了就是非常的肥胖。
"""
if a<18.5:
return 0
elif 18.5<=a<=24:
return 1
elif 24<a<=27:
return 2
elif 27<a<=32:
return 3
else:
return 4
data['BMI']=data['体重指数'].apply(BMI)
def DBP(a):
"""
舒张压范围为60-90
"""
if a<60:
return 0
elif 60<=a<=90:
return 1
elif a>90:
return 2
else:
return a
data['DBP']=data['舒张压'].apply(DBP)
for col in ['糖尿病家族史','性别']:
data[col] = data[col].astype('category')
print(data.shape)
训练集20%划分为验证集
feature_name = [i for i in data.columns if i not in ['编号', '患有糖尿病标识']]
# cat_list = [i for i in train.columns if i not in ['编号', '患有糖尿病标识']] # categorical_feature 专业写法
X_train = data[data['患有糖尿病标识']!=-1][list(set(feature_name))].reset_index(drop=True)
y = data[data['患有糖尿病标识']!=-1]['患有糖尿病标识'].reset_index(drop=True).astype(int)
X_test = data[data['患有糖尿病标识']==-1][list(set(feature_name))].reset_index(drop=True)
print('X_train',X_train.shape,'X_test',X_test.shape)
print('\n训练集20%划分为验证集')
train_X,val_X,train_y,val_y = train_test_split(X_train,y,test_size=0.3,random_state=5)
print('train_X',train_X.shape,'val_X',val_X.shape)
lr = LR(C=1e2,random_state=2020)
lr.fit(train_X,train_y)
prediction_lr = lr.predict(val_X)
print('\nF1 score:',f1_score(val_y,prediction_lr))
X_train (5070, 12) X_test (1000, 12)
# 训练集20%划分为验证集
train_X (3549, 12) val_X (1521, 12)
F1 score: 0.721554116558742
Onehot Encoding
pd.get_dummies(data_train,columns=['糖尿病家族史'])
from sklearn.preprocessing import OneHotEncoder
one_hot = OneHotEncoder()
one_hot.fit_transform(data_train[['糖尿病家族史']]).toarray()
Label Encoding
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
data_train['糖尿病家族史_LabelEncoder'] = le.fit_transform(data_train['糖尿病家族史'])
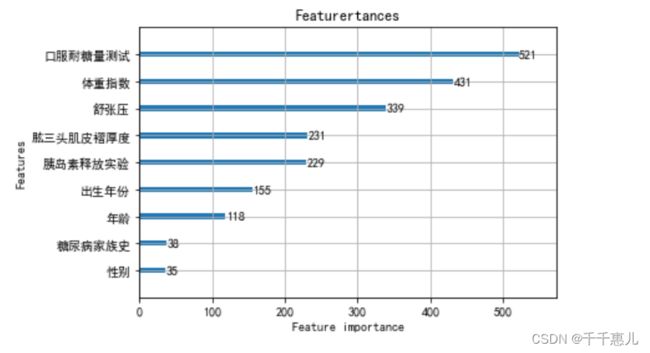
Task5:特征筛选
思路:
1、根据特征之间的相关性,剔除与标签相关性低的特征
2、计算特征重要性,剔除特征贡献度低的特征
3、其它方法
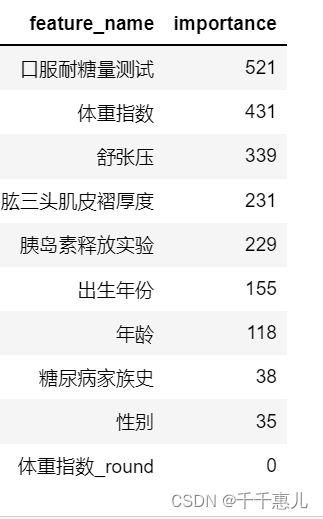
importance = clf.feature_importances_
feature_name = clf.feature_name_
feature_importance = pd.DataFrame({'feature_name':feature_name,'importance':importance} )
feature_importance = feature_importance.sort_values(by=['importance'],ascending=False)
feature_importance.to_csv('feature_importance.csv',index=False)
import matplotlib.pyplot as plt
plt.figure(figsize=(24,12))
lgb.plot_importance(clf, max_num_features=15)
plt.title("Featurertances")
plt.show()
使用树模型完成模型的训练,通过特征重要性筛选出Top5的特征;使用筛选出的特征和逻辑回归进行训练,验证集精度由0.72155提高到了 0.72910。
fea_top5 = ['口服耐糖量测试','体重指数','舒张压','肱三头肌皮褶厚度','胰岛素释放实验']
X_train = data[data['患有糖尿病标识']!=-1][list(set(fea_top5))].reset_index(drop=True)
y = data[data['患有糖尿病标识']!=-1]['患有糖尿病标识'].reset_index(drop=True).astype(int)
X_test = data[data['患有糖尿病标识']==-1][list(set(fea_top5))].reset_index(drop=True)
print('X_train',X_train.shape,'X_test',X_test.shape)
print('\n训练集20%划分为验证集')
train_X,val_X,train_y,val_y = train_test_split(X_train,y,test_size=0.3,random_state=5)
print('train_X',train_X.shape,'val_X',val_X.shape)
lr = LR(C=1e2,random_state=2020)
lr.fit(train_X,train_y)
prediction_lr = lr.predict(val_X)
print('\nF1 score:',f1_score(val_y,prediction_lr))
X_train (5070, 5) X_test (1000, 5)
训练集20%划分为验证集
train_X (3549, 5) val_X (1521, 5)
F1 score: 0.72910927456382
Task6:高阶树模型
import lightgbm as lgb
# 模型交叉验证
def run_model_cv(model, kf, X_tr, y, X_te, cate_col=None):
train_pred = np.zeros( (len(X_tr), len(np.unique(y))) )
test_pred = np.zeros( (len(X_te), len(np.unique(y))) )
cv_clf = []
for tr_idx, val_idx in kf.split(X_tr, y):
x_tr = X_tr.iloc[tr_idx]; y_tr = y.iloc[tr_idx]
x_val = X_tr.iloc[val_idx]; y_val = y.iloc[val_idx]
call_back = [
lgb.early_stopping(50),
]
eval_set = [(x_val, y_val)]
model.fit(x_tr, y_tr, eval_set=eval_set, callbacks=call_back, verbose=-1)
cv_clf.append(model)
train_pred[val_idx] = model.predict_proba(x_val)
test_pred += model.predict_proba(X_te)
test_pred /= kf.n_splits
return train_pred, test_pred, cv_clf
clf = lgb.LGBMClassifier(
max_depth=3,
n_estimators=4000,
n_jobs=-1,
verbose=-1,
verbosity=-1,
learning_rate=0.1,
)
train_pred, test_pred, cv_clf = run_model_cv(
clf, KFold(n_splits=5),
df_train.drop(['编号', '患有糖尿病标识'], axis=1),
df_train['患有糖尿病标识'],
df_test.drop(['编号', '患有糖尿病标识'], axis=1),
)
print((train_pred.argmax(1) == df_test['患有糖尿病标识']).mean())
df_test['label'] = test_pred.argmax(1)
df_test.rename({'编号': 'uuid'}, axis=1)[['uuid', 'label']].to_csv('submit_lgb_0628.csv', index=None)
Task7:多折训练与集成
params = {
'num_leaves': 31, #叶子节点数,默认31,树模型复杂度 num_leaves<2^max_depth
'min_data_in_leaf': 15, # 也叫min_child_samples,将其设置的较大可以避免生成一个过深的树, 但有可能导致欠拟合。
'objective': 'binary', # 目标函数,可选 ‘regression’ binary 二分类(二进制) multiclass: 多分类
'learning_rate': 0.03, # 学习率 默认 0.1 取值范围:0.01-0.3 学习率越小越好
"boosting": "gbdt", # 提升类型,默认 gbdt
"feature_fraction": 0.78, # 建树的特征选择比例 ,boosting为 rf 时用 在不进行重采样的情况下随机选择部分数据
"bagging_fraction": 0.85, # 建树的样本采样比例,用于加快训练速度、防止过拟合
'bagging_freq':3, # 3或5 表示bagging的频率,每 k 次迭代执行bagging ( bagging_fraction + bagging_freq 必须同时设置)
"bagging_seed": 1017,
"metric": 'auc', # 评估函数 可选 'auc','mae','mse','rmse','binary_logloss','multi_logloss','l2'
"lambda_l1": 0.1, # L1正则化,默认为0
"nthread": -1, # lgb线程数,一般设置为 CPU的内核数, -1 是自动匹配
'is_unbalance':True, # 用于 binary 分类 ,如果培训数据不平衡 设置为 true
# 'device ': 'gpu',
# 'gpu_platform_id ': 0,
# 'gpu_device_id' : 0
}
print('Start training...')
for i, (train_index, test_index) in enumerate(skf.split(X_train, y), start=1):
print(i)
trn_data = lgb.Dataset(X_train[feature_name].iloc[train_index], label=y.iloc[train_index])
# 加载验证集
val_data = lgb.Dataset(X_train[feature_name].iloc[test_index], label=y.iloc[test_index])
# 迭代次数
num_round = 10000
clf = lgb.train(params, trn_data, num_round, feval=f1_score_vail,valid_sets = [trn_data, val_data],
verbose_eval= 100, early_stopping_rounds = 200)
cv_model.append(clf)
clf.save_model('lgb_model.txt')
y_test = clf.predict(X_test[feature_name])
y_val = clf.predict(X_train[feature_name].iloc[test_index], num_iteration=clf.best_iteration)
oof_lgb[test_index] = y_val
cv_score.append(f1_score(y.iloc[test_index],(y_val >=0.5).astype(int)))
Start training...
1
[LightGBM] [Info] Number of positive: 1549, number of negative: 2507
[LightGBM] [Warning] Auto-choosing col-wise multi-threading, the overhead of testing was 0.000687 seconds.
You can set `force_col_wise=true` to remove the overhead.
[LightGBM] [Info] Total Bins 1091
[LightGBM] [Info] Number of data points in the train set: 4056, number of used features: 10
[LightGBM] [Info] [binary:BoostFromScore]: pavg=0.381903 -> initscore=-0.481477
[LightGBM] [Info] Start training from score -0.481477
Training until validation scores don't improve for 200 rounds
[100] training's auc: 0.997866 training's f1_score: 0.966921 valid_1's auc: 0.989895 valid_1's f1_score: 0.926209
[200] training's auc: 0.999613 training's f1_score: 0.982748 valid_1's auc: 0.990942 valid_1's f1_score: 0.927757
Early stopping, best iteration is:
[49] training's auc: 0.996568 training's f1_score: 0.962584 valid_1's auc: 0.988172 valid_1's f1_score: 0.932312
……
5
[LightGBM] [Info] Number of positive: 1548, number of negative: 2508
[LightGBM] [Warning] Auto-choosing row-wise multi-threading, the overhead of testing was 0.000219 seconds.
You can set `force_row_wise=true` to remove the overhead.
And if memory is not enough, you can set `force_col_wise=true`.
[LightGBM] [Info] Total Bins 1096
[LightGBM] [Info] Number of data points in the train set: 4056, number of used features: 10
[LightGBM] [Info] [binary:BoostFromScore]: pavg=0.381657 -> initscore=-0.482522
[LightGBM] [Info] Start training from score -0.482522
Training until validation scores don't improve for 200 rounds
[100] training's auc: 0.997908 training's f1_score: 0.967329 valid_1's auc: 0.991967 valid_1's f1_score: 0.950695
[200] training's auc: 0.999629 training's f1_score: 0.986231 valid_1's auc: 0.992375 valid_1's f1_score: 0.946835
Early stopping, best iteration is:
[5] training's auc: 0.993822 training's f1_score: 0 valid_1's auc: 0.994075 valid_1's f1_score: 0