sklearn常用分类算法分析乳腺癌数据
模型评估
参考网址:机器学习分类模型评价指标详述 - 知乎
机器学习模型评估的方法总结(回归、分类模型的评估)_人工智能_sinat_16388393的博客-CSDN博客
【机器学习】Sklearn 常用分类器(全)_人工智能_白糖炒栗子-CSDN博客
python-sklearn常用分类算法模型的调用_python_脚本之家
机器学习各种算法怎么调参?—知乎
GBDT、XGBoost、LightGBM-的使用及参数调优—简书
和鲸社区 - Kesci.com
1. 机器学习常用分类模型:
1.最近邻 (KNN Classifier)
2.Logistic回归 (Logistic Regression Classifier)
3.高斯朴素贝叶斯(GaussianNB)
4.多项分布朴素贝叶斯(Multinomial Naive Bayes Classifier )
5.决策树(Decision Tree Classifier)
6.集成算法(Ensemble methods)
- 梯度提升决策树(GBDT(Gradient Boosting Decision Tree) Classifier)
- 自适应推举算法(AdaBoost)(AdaBoost Classifier)
- 随机森林 (Random Forest Classifier)
- Bagging
7.支持向量机(SVM Classifier)
2.分类模型的评估:
-
模型评估指标
准确率,精确率和召回率,F1分数,均方误差、根均方误差、绝对百分比误差,ROC曲线 -
模型评估方法
Holdout检验,交叉验证,自助法,超参数调优 -
优化过拟合与欠拟合
- 降低过拟合风险的方法:
(1). 从数据入手,获得更多的训练数据。使用更多的训练数据是解决过拟合问题最有效的手段,因为更多的样本能够让模型学习到更多更有效的特征,减少噪音的影响,当然,直接增加实验数据一般是很困难的,但是可以通过一定的规则来扩充训练数据。比如,在图像分类的问题上,可以通过图像的平移、旋转、缩放等方式扩充数据;更进一步地,可以使用生成式对抗网络来合成大量的新训练数据
(2). 降低模型复杂度。在数据较少时,模型过于复杂是产生过拟合的主要因素,适当降低模型复杂度可以避免拟合过多的采样噪音。例如,在神经网络中减少网络层数、神经元个数等;在决策树模型中降低树的深度、进行剪枝等
(3). 正则化方法
(4). 集成学习方法。集成学习是把多个模型集成在一起,来降低单一模型的过拟合风险
- 降低欠拟合风险方法
(1). 添加新特征。当特征不足或现有特征与样本标签的相关性不强时,模型容易出现不拟合,通过挖掘’上下文特征’‘ID类特征’'组合特征’等新的特征,往往能够取得更好的效果,在深度学习的潮流中,有很多类型可以帮组完成特征工程,如因子分解机
(2). 增加模型复杂度。简单模型的学习能力较差,通过增加模型的复杂度可以使模型拥有更强的拟合能力,例如,在线性模型中添加高次项,在神经网络模型中增加网络层数或神经元个数等
(3). 减少正则化系数。正则化是用来防止过拟合的,但当模型出现欠拟合现象时,则需要针对性地减少正则化系数
1. 导入扩展库
import time
from sklearn import metrics
import pickle as pickle
import pandas as pd
from sklearn import tree
from sklearn.tree import export_graphviz
import graphviz
from IPython.display import Image
import pydotplus
import os
from sklearn.datasets import load_breast_cancer
from sklearn.model_selection import train_test_split
from sklearn.naive_bayes import MultinomialNB
from sklearn.naive_bayes import GaussianNB
from sklearn.neighbors import KNeighborsClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.svm import SVC
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import AdaBoostClassifier
from sklearn.ensemble import BaggingClassifier
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import learning_curve
from common.utils import plot_learning_curve
from common.utils import plot_param_curve
from sklearn.metrics import roc_curve, auc
from sklearn.metrics import plot_roc_curve
from sklearn.metrics import confusion_matrix
from sklearn.metrics import classification_report
from sklearn.model_selection import ShuffleSplit
import numpy as np
import matplotlib.pyplot as plt
import matplotlib as mpl
mpl.rcParams['font.sans-serif'] = ['SimHei'] #设置简黑字体
mpl.rcParams['axes.unicode_minus'] = False # 解决‘-’bug
%matplotlib inline
import warnings
warnings.filterwarnings("ignore")
2. 准备训练数据
cancer = load_breast_cancer() #载入数据
df = pd.DataFrame(cancer.data,columns=cancer.feature_names)
df['target'] = cancer.target
x = cancer.data
y = cancer.target
print('data:',x.shape)
print('target:',y.shape)
# 打印前五行数据
df.head()
data: (569, 30)
target: (569,)
| mean radius | mean texture | mean perimeter | mean area | mean smoothness | mean compactness | mean concavity | mean concave points | mean symmetry | mean fractal dimension | ... | worst texture | worst perimeter | worst area | worst smoothness | worst compactness | worst concavity | worst concave points | worst symmetry | worst fractal dimension | target | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 17.99 | 10.38 | 122.80 | 1001.0 | 0.11840 | 0.27760 | 0.3001 | 0.14710 | 0.2419 | 0.07871 | ... | 17.33 | 184.60 | 2019.0 | 0.1622 | 0.6656 | 0.7119 | 0.2654 | 0.4601 | 0.11890 | 0 |
| 1 | 20.57 | 17.77 | 132.90 | 1326.0 | 0.08474 | 0.07864 | 0.0869 | 0.07017 | 0.1812 | 0.05667 | ... | 23.41 | 158.80 | 1956.0 | 0.1238 | 0.1866 | 0.2416 | 0.1860 | 0.2750 | 0.08902 | 0 |
| 2 | 19.69 | 21.25 | 130.00 | 1203.0 | 0.10960 | 0.15990 | 0.1974 | 0.12790 | 0.2069 | 0.05999 | ... | 25.53 | 152.50 | 1709.0 | 0.1444 | 0.4245 | 0.4504 | 0.2430 | 0.3613 | 0.08758 | 0 |
| 3 | 11.42 | 20.38 | 77.58 | 386.1 | 0.14250 | 0.28390 | 0.2414 | 0.10520 | 0.2597 | 0.09744 | ... | 26.50 | 98.87 | 567.7 | 0.2098 | 0.8663 | 0.6869 | 0.2575 | 0.6638 | 0.17300 | 0 |
| 4 | 20.29 | 14.34 | 135.10 | 1297.0 | 0.10030 | 0.13280 | 0.1980 | 0.10430 | 0.1809 | 0.05883 | ... | 16.67 | 152.20 | 1575.0 | 0.1374 | 0.2050 | 0.4000 | 0.1625 | 0.2364 | 0.07678 | 0 |
5 rows × 31 columns
# 查看数据描述
df.info()
RangeIndex: 569 entries, 0 to 568
Data columns (total 31 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 mean radius 569 non-null float64
1 mean texture 569 non-null float64
2 mean perimeter 569 non-null float64
3 mean area 569 non-null float64
4 mean smoothness 569 non-null float64
5 mean compactness 569 non-null float64
6 mean concavity 569 non-null float64
7 mean concave points 569 non-null float64
8 mean symmetry 569 non-null float64
9 mean fractal dimension 569 non-null float64
10 radius error 569 non-null float64
11 texture error 569 non-null float64
12 perimeter error 569 non-null float64
13 area error 569 non-null float64
14 smoothness error 569 non-null float64
15 compactness error 569 non-null float64
16 concavity error 569 non-null float64
17 concave points error 569 non-null float64
18 symmetry error 569 non-null float64
19 fractal dimension error 569 non-null float64
20 worst radius 569 non-null float64
21 worst texture 569 non-null float64
22 worst perimeter 569 non-null float64
23 worst area 569 non-null float64
24 worst smoothness 569 non-null float64
25 worst compactness 569 non-null float64
26 worst concavity 569 non-null float64
27 worst concave points 569 non-null float64
28 worst symmetry 569 non-null float64
29 worst fractal dimension 569 non-null float64
30 target 569 non-null int32
dtypes: float64(30), int32(1)
memory usage: 135.7 KB
数据未包含空值
# 打印数据类别及每种类别的个数
df['target'].value_counts()
1 357
0 212
Name: target, dtype: int64
# 查看对数值属性的概括
df.describe()
| mean radius | mean texture | mean perimeter | mean area | mean smoothness | mean compactness | mean concavity | mean concave points | mean symmetry | mean fractal dimension | ... | worst texture | worst perimeter | worst area | worst smoothness | worst compactness | worst concavity | worst concave points | worst symmetry | worst fractal dimension | target | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | ... | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 |
| mean | 14.127292 | 19.289649 | 91.969033 | 654.889104 | 0.096360 | 0.104341 | 0.088799 | 0.048919 | 0.181162 | 0.062798 | ... | 25.677223 | 107.261213 | 880.583128 | 0.132369 | 0.254265 | 0.272188 | 0.114606 | 0.290076 | 0.083946 | 0.627417 |
| std | 3.524049 | 4.301036 | 24.298981 | 351.914129 | 0.014064 | 0.052813 | 0.079720 | 0.038803 | 0.027414 | 0.007060 | ... | 6.146258 | 33.602542 | 569.356993 | 0.022832 | 0.157336 | 0.208624 | 0.065732 | 0.061867 | 0.018061 | 0.483918 |
| min | 6.981000 | 9.710000 | 43.790000 | 143.500000 | 0.052630 | 0.019380 | 0.000000 | 0.000000 | 0.106000 | 0.049960 | ... | 12.020000 | 50.410000 | 185.200000 | 0.071170 | 0.027290 | 0.000000 | 0.000000 | 0.156500 | 0.055040 | 0.000000 |
| 25% | 11.700000 | 16.170000 | 75.170000 | 420.300000 | 0.086370 | 0.064920 | 0.029560 | 0.020310 | 0.161900 | 0.057700 | ... | 21.080000 | 84.110000 | 515.300000 | 0.116600 | 0.147200 | 0.114500 | 0.064930 | 0.250400 | 0.071460 | 0.000000 |
| 50% | 13.370000 | 18.840000 | 86.240000 | 551.100000 | 0.095870 | 0.092630 | 0.061540 | 0.033500 | 0.179200 | 0.061540 | ... | 25.410000 | 97.660000 | 686.500000 | 0.131300 | 0.211900 | 0.226700 | 0.099930 | 0.282200 | 0.080040 | 1.000000 |
| 75% | 15.780000 | 21.800000 | 104.100000 | 782.700000 | 0.105300 | 0.130400 | 0.130700 | 0.074000 | 0.195700 | 0.066120 | ... | 29.720000 | 125.400000 | 1084.000000 | 0.146000 | 0.339100 | 0.382900 | 0.161400 | 0.317900 | 0.092080 | 1.000000 |
| max | 28.110000 | 39.280000 | 188.500000 | 2501.000000 | 0.163400 | 0.345400 | 0.426800 | 0.201200 | 0.304000 | 0.097440 | ... | 49.540000 | 251.200000 | 4254.000000 | 0.222600 | 1.058000 | 1.252000 | 0.291000 | 0.663800 | 0.207500 | 1.000000 |
8 rows × 31 columns
# 画出数据分布直方图
df.hist(bins=50,figsize=(20,15))
array([[,
,
,
,
,
],
[,
,
,
,
,
],
[,
,
,
,
,
],
[,
,
,
,
,
],
[,
,
,
,
,
],
[,
,
,
,
,
]],
dtype=object)
x_train,x_test,y_train,y_test = train_test_split(x,y,test_size=0.33)
训练集
df_train = pd.DataFrame(x_train,columns=cancer.feature_names)
df_train['target'] = y_train
df_train
| mean radius | mean texture | mean perimeter | mean area | mean smoothness | mean compactness | mean concavity | mean concave points | mean symmetry | mean fractal dimension | ... | worst texture | worst perimeter | worst area | worst smoothness | worst compactness | worst concavity | worst concave points | worst symmetry | worst fractal dimension | target | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 20.510 | 27.81 | 134.40 | 1319.0 | 0.09159 | 0.10740 | 0.15540 | 0.08340 | 0.1448 | 0.05592 | ... | 37.38 | 162.70 | 1872.0 | 0.1223 | 0.27610 | 0.4146 | 0.15630 | 0.2437 | 0.08328 | 0 |
| 1 | 15.750 | 20.25 | 102.60 | 761.3 | 0.10250 | 0.12040 | 0.11470 | 0.06462 | 0.1935 | 0.06303 | ... | 30.29 | 125.90 | 1088.0 | 0.1552 | 0.44800 | 0.3976 | 0.14790 | 0.3993 | 0.10640 | 0 |
| 2 | 12.470 | 17.31 | 80.45 | 480.1 | 0.08928 | 0.07630 | 0.03609 | 0.02369 | 0.1526 | 0.06046 | ... | 24.34 | 92.82 | 607.3 | 0.1276 | 0.25060 | 0.2028 | 0.10530 | 0.3035 | 0.07661 | 1 |
| 3 | 13.660 | 19.13 | 89.46 | 575.3 | 0.09057 | 0.11470 | 0.09657 | 0.04812 | 0.1848 | 0.06181 | ... | 25.50 | 101.40 | 708.8 | 0.1147 | 0.31670 | 0.3660 | 0.14070 | 0.2744 | 0.08839 | 1 |
| 4 | 8.888 | 14.64 | 58.79 | 244.0 | 0.09783 | 0.15310 | 0.08606 | 0.02872 | 0.1902 | 0.08980 | ... | 15.67 | 62.56 | 284.4 | 0.1207 | 0.24360 | 0.1434 | 0.04786 | 0.2254 | 0.10840 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 376 | 8.219 | 20.70 | 53.27 | 203.9 | 0.09405 | 0.13050 | 0.13210 | 0.02168 | 0.2222 | 0.08261 | ... | 29.72 | 58.08 | 249.8 | 0.1630 | 0.43100 | 0.5381 | 0.07879 | 0.3322 | 0.14860 | 1 |
| 377 | 15.460 | 19.48 | 101.70 | 748.9 | 0.10920 | 0.12230 | 0.14660 | 0.08087 | 0.1931 | 0.05796 | ... | 26.00 | 124.90 | 1156.0 | 0.1546 | 0.23940 | 0.3791 | 0.15140 | 0.2837 | 0.08019 | 0 |
| 378 | 12.780 | 16.49 | 81.37 | 502.5 | 0.09831 | 0.05234 | 0.03653 | 0.02864 | 0.1590 | 0.05653 | ... | 19.76 | 85.67 | 554.9 | 0.1296 | 0.07061 | 0.1039 | 0.05882 | 0.2383 | 0.06410 | 1 |
| 379 | 20.730 | 31.12 | 135.70 | 1419.0 | 0.09469 | 0.11430 | 0.13670 | 0.08646 | 0.1769 | 0.05674 | ... | 47.16 | 214.00 | 3432.0 | 0.1401 | 0.26440 | 0.3442 | 0.16590 | 0.2868 | 0.08218 | 0 |
| 380 | 10.860 | 21.48 | 68.51 | 360.5 | 0.07431 | 0.04227 | 0.00000 | 0.00000 | 0.1661 | 0.05948 | ... | 24.77 | 74.08 | 412.3 | 0.1001 | 0.07348 | 0.0000 | 0.00000 | 0.2458 | 0.06592 | 1 |
381 rows × 31 columns
测试集
df_test = pd.DataFrame(x_test,columns=cancer.feature_names)
df_test['target'] = y_test
df_test
| mean radius | mean texture | mean perimeter | mean area | mean smoothness | mean compactness | mean concavity | mean concave points | mean symmetry | mean fractal dimension | ... | worst texture | worst perimeter | worst area | worst smoothness | worst compactness | worst concavity | worst concave points | worst symmetry | worst fractal dimension | target | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 20.48 | 21.46 | 132.50 | 1306.0 | 0.08355 | 0.08348 | 0.09042 | 0.06022 | 0.1467 | 0.05177 | ... | 26.17 | 161.70 | 1750.0 | 0.1228 | 0.2311 | 0.3158 | 0.14450 | 0.2238 | 0.07127 | 0 |
| 1 | 14.96 | 19.10 | 97.03 | 687.3 | 0.08992 | 0.09823 | 0.05940 | 0.04819 | 0.1879 | 0.05852 | ... | 26.19 | 109.10 | 809.8 | 0.1313 | 0.3030 | 0.1804 | 0.14890 | 0.2962 | 0.08472 | 1 |
| 2 | 19.53 | 18.90 | 129.50 | 1217.0 | 0.11500 | 0.16420 | 0.21970 | 0.10620 | 0.1792 | 0.06552 | ... | 26.24 | 171.10 | 2053.0 | 0.1495 | 0.4116 | 0.6121 | 0.19800 | 0.2968 | 0.09929 | 0 |
| 3 | 12.40 | 17.68 | 81.47 | 467.8 | 0.10540 | 0.13160 | 0.07741 | 0.02799 | 0.1811 | 0.07102 | ... | 22.91 | 89.61 | 515.8 | 0.1450 | 0.2629 | 0.2403 | 0.07370 | 0.2556 | 0.09359 | 1 |
| 4 | 12.16 | 18.03 | 78.29 | 455.3 | 0.09087 | 0.07838 | 0.02916 | 0.01527 | 0.1464 | 0.06284 | ... | 27.87 | 88.83 | 547.4 | 0.1208 | 0.2279 | 0.1620 | 0.05690 | 0.2406 | 0.07729 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 183 | 11.93 | 21.53 | 76.53 | 438.6 | 0.09768 | 0.07849 | 0.03328 | 0.02008 | 0.1688 | 0.06194 | ... | 26.15 | 87.54 | 583.0 | 0.1500 | 0.2399 | 0.1503 | 0.07247 | 0.2438 | 0.08541 | 1 |
| 184 | 12.23 | 19.56 | 78.54 | 461.0 | 0.09586 | 0.08087 | 0.04187 | 0.04107 | 0.1979 | 0.06013 | ... | 28.36 | 92.15 | 638.4 | 0.1429 | 0.2042 | 0.1377 | 0.10800 | 0.2668 | 0.08174 | 1 |
| 185 | 11.04 | 14.93 | 70.67 | 372.7 | 0.07987 | 0.07079 | 0.03546 | 0.02074 | 0.2003 | 0.06246 | ... | 20.83 | 79.73 | 447.1 | 0.1095 | 0.1982 | 0.1553 | 0.06754 | 0.3202 | 0.07287 | 1 |
| 186 | 13.70 | 17.64 | 87.76 | 571.1 | 0.09950 | 0.07957 | 0.04548 | 0.03160 | 0.1732 | 0.06088 | ... | 23.53 | 95.78 | 686.5 | 0.1199 | 0.1346 | 0.1742 | 0.09077 | 0.2518 | 0.06960 | 1 |
| 187 | 19.07 | 24.81 | 128.30 | 1104.0 | 0.09081 | 0.21900 | 0.21070 | 0.09961 | 0.2310 | 0.06343 | ... | 33.17 | 177.40 | 1651.0 | 0.1247 | 0.7444 | 0.7242 | 0.24930 | 0.4670 | 0.10380 | 0 |
188 rows × 31 columns
3.创建模型
# Multinomial Naive Bayes Classifier
def mul_naive_bayes_classifier(train_x, train_y):
model = MultinomialNB(alpha=0.01)
model.fit(train_x, train_y)
return model
def naive_bayes_classifier(train_x, train_y):
model = GaussianNB(priors=None)
model.fit(train_x, train_y)
return model
# KNN Classifier
def knn_classifier(train_x, train_y):
model = KNeighborsClassifier()
model.fit(train_x, train_y)
return model
# Logistic Regression Classifier
def logistic_regression_classifier(train_x, train_y):
model = LogisticRegression(penalty='l2')
model.fit(train_x, train_y)
return model
# Random Forest Classifier
def random_forest_classifier(train_x, train_y):
model = RandomForestClassifier(n_estimators=8)
model.fit(train_x, train_y)
return model
# Decision Tree Classifier
def decision_tree_classifier(train_x, train_y):
model = DecisionTreeClassifier()
model.fit(train_x, train_y)
return model
# GBDT(Gradient Boosting Decision Tree) Classifier
def gradient_boosting_classifier(train_x, train_y):
model = GradientBoostingClassifier(n_estimators=200)
model.fit(train_x, train_y)
return model
# SVM Classifier
def svm_classifier(train_x, train_y):
model = SVC(kernel='rbf', probability=True)
model.fit(train_x, train_y)
return model
def adaboost_classifier(train_x, train_y):
model = AdaBoostClassifier(DecisionTreeClassifier(),algorithm="SAMME", n_estimators=7, learning_rate=0.4)
model.fit(train_x, train_y)
return model
def bagging_classifier(train_x, train_y):
model = BaggingClassifier(DecisionTreeClassifier(), bootstrap=True)
model.fit(train_x,train_y)
return model
4.测试模型
test_classifiers = ['NB(高斯朴素贝叶斯)','MNB(多项式分布朴素贝叶斯)', 'KNN(最近邻)', 'LR(Logistic回归)', 'RF(随机森林)', 'DT(决策树)', 'SVM(支持向量机)', 'GBDT(梯度提升决策树)','Adaboost','Bagging']
classifiers = {
'GBDT(梯度提升决策树)':gradient_boosting_classifier,
'Adaboost':adaboost_classifier,
'Bagging':bagging_classifier,
'NB(高斯朴素贝叶斯)':naive_bayes_classifier,
'MNB(多项式分布朴素贝叶斯)':mul_naive_bayes_classifier,
'KNN(最近邻)':knn_classifier,
'LR(Logistic回归)':logistic_regression_classifier,
'RF(随机森林)':random_forest_classifier,
'DT(决策树)':decision_tree_classifier,
'SVM(支持向量机)':svm_classifier
}
for classifier in test_classifiers:
print('******************* %s ********************' % classifier)
start_time = time.time()
model = classifiers[classifier](x_train, y_train)
print(model)
print('training took %fs!' % (time.time() - start_time))
predict = model.predict(x_test)
# if model_save_file != None:
# model_save[classifier] = model )
score = metrics.precision_score(y_test, predict)
recall = metrics.recall_score(y_test, predict)
print('precision: %.2f%%, recall: %.2f%%' % (100 * score, 100 * recall))
accuracy = metrics.accuracy_score(y_test, predict)
print('accuracy: %.2f%%' % (100 * accuracy))
c_matrix = confusion_matrix(
y_test, # array, Gound true (correct) target values
predict, # array, Estimated targets as returned by a classifier
labels=[0,1], # array, List of labels to index the matrix.
sample_weight=None # array-like of shape = [n_samples], Optional sample weights
)
print('\nclassification_report:')
print(classification_report( y_test,predict,labels=[0,1]))
print('\nconfusion_matrix:')
print(c_matrix)
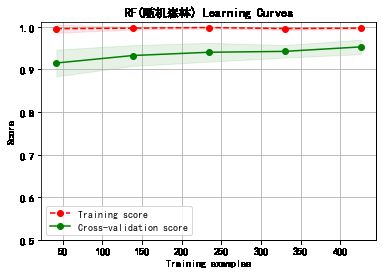
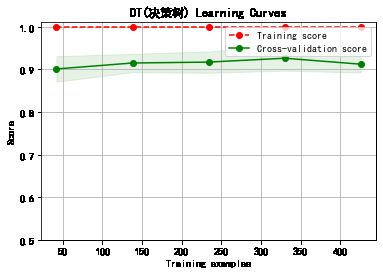
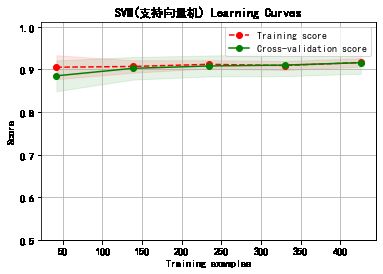
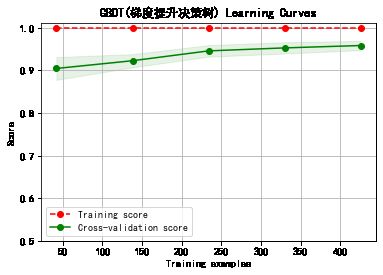
cv = ShuffleSplit(n_splits=10, test_size=0.25, random_state=0)
title = classifier+' Learning Curves'
start = time.clock()
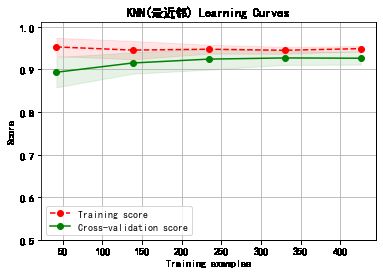
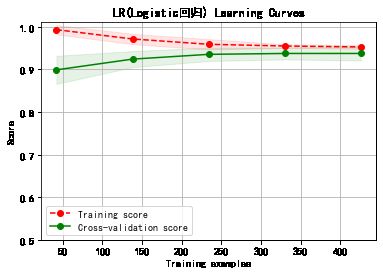
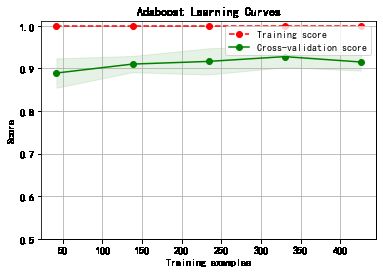
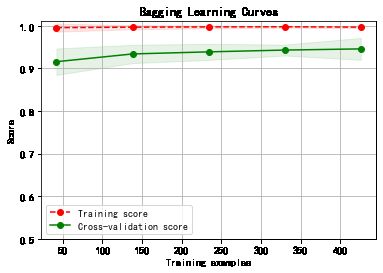
plot_learning_curve(plt, model,title,cancer.data, cancer.target, ylim=(0.5, 1.01), cv=cv)
print('elaspe: {0:.6f}'.format(time.clock()-start))
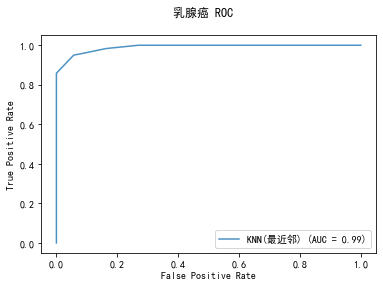
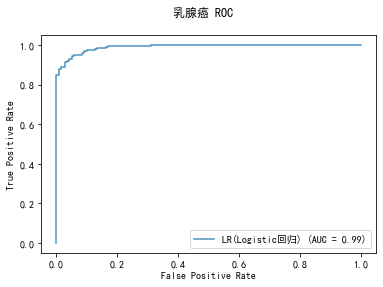
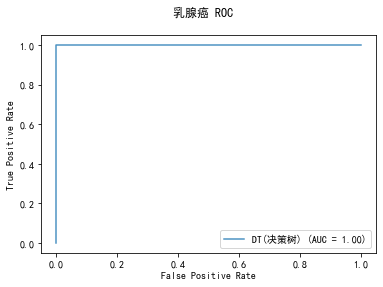
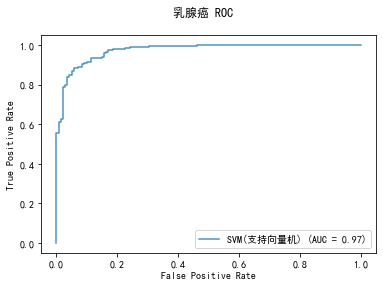
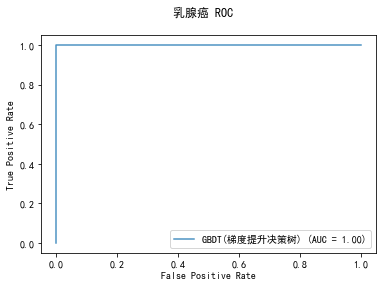
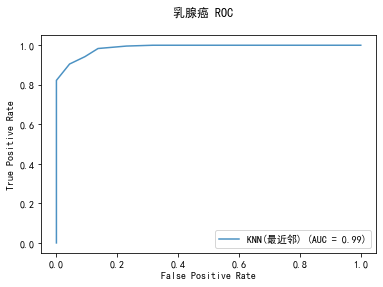
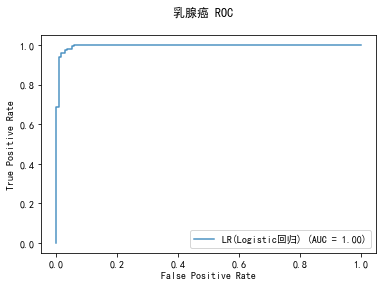
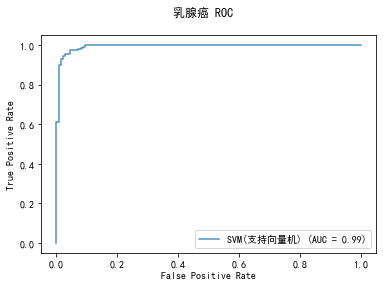
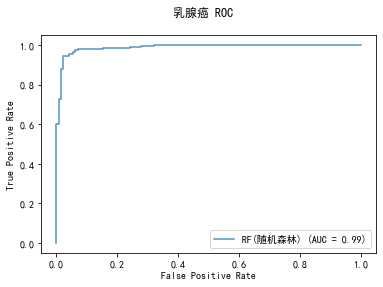
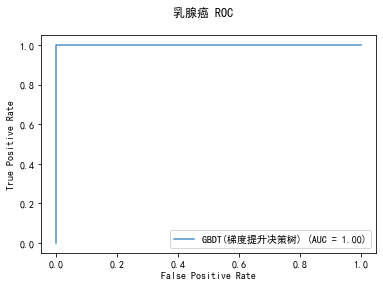
curve1 = plot_roc_curve(model, x_train, y_train, alpha=0.8,name=classifier)
curve1.figure_.suptitle("乳腺癌 ROC")
#画出决策树
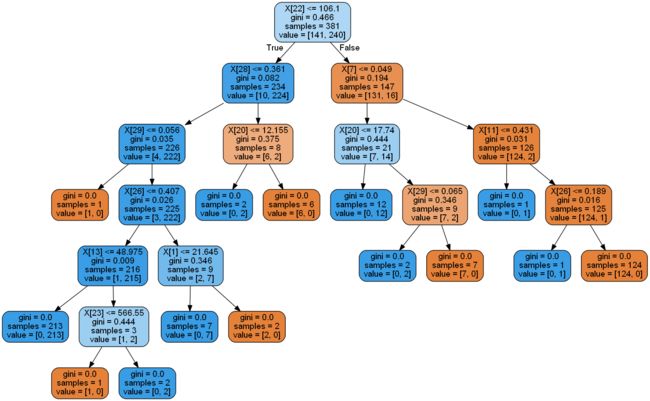
if classifier == 'DT(决策树)':
dot_data = export_graphviz(model,
out_file = None,
# feature_names = iris_feature_name,
# class_names = iris_target_name,
filled=True,
rounded=True
)
graph = pydotplus.graph_from_dot_data(dot_data)
display(Image(graph.create_png()))
plt.show()
print()
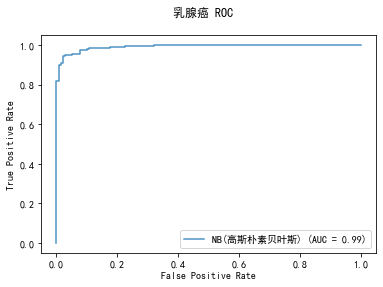
******************* NB(高斯朴素贝叶斯) ********************
GaussianNB(priors=None, var_smoothing=1e-09)
training took 0.004002s!
precision: 91.67%, recall: 94.02%
accuracy: 90.96%
classification_report:
precision recall f1-score support
0 0.90 0.86 0.88 71
1 0.92 0.94 0.93 117
accuracy 0.91 188
macro avg 0.91 0.90 0.90 188
weighted avg 0.91 0.91 0.91 188
confusion_matrix:
[[ 61 10]
[ 7 110]]
elaspe: 0.299883
![]()
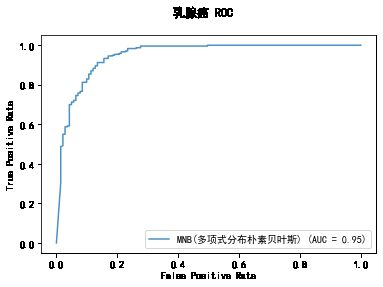
******************* MNB(多项式分布朴素贝叶斯) ********************
MultinomialNB(alpha=0.01, class_prior=None, fit_prior=True)
training took 0.008931s!
precision: 88.19%, recall: 95.73%
accuracy: 89.36%
classification_report:
precision recall f1-score support
0 0.92 0.79 0.85 71
1 0.88 0.96 0.92 117
accuracy 0.89 188
macro avg 0.90 0.87 0.88 188
weighted avg 0.90 0.89 0.89 188
confusion_matrix:
[[ 56 15]
[ 5 112]]
elaspe: 0.272553
******************* KNN(最近邻) ********************
KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski',
metric_params=None, n_jobs=None, n_neighbors=5, p=2,
weights='uniform')
training took 0.006923s!
precision: 93.28%, recall: 94.87%
accuracy: 92.55%
classification_report:
precision recall f1-score support
0 0.91 0.89 0.90 71
1 0.93 0.95 0.94 117
accuracy 0.93 188
macro avg 0.92 0.92 0.92 188
weighted avg 0.93 0.93 0.93 188
confusion_matrix:
[[ 63 8]
[ 6 111]]
elaspe: 1.937058
******************* LR(Logistic回归) ********************
LogisticRegression(C=1.0, class_weight=None, dual=False, fit_intercept=True,
intercept_scaling=1, l1_ratio=None, max_iter=100,
multi_class='auto', n_jobs=None, penalty='l2',
random_state=None, solver='lbfgs', tol=0.0001, verbose=0,
warm_start=False)
training took 0.132035s!
precision: 95.73%, recall: 95.73%
accuracy: 94.68%
classification_report:
precision recall f1-score support
0 0.93 0.93 0.93 71
1 0.96 0.96 0.96 117
accuracy 0.95 188
macro avg 0.94 0.94 0.94 188
weighted avg 0.95 0.95 0.95 188
confusion_matrix:
[[ 66 5]
[ 5 112]]
elaspe: 5.063377
******************* RF(随机森林) ********************
RandomForestClassifier(bootstrap=True, ccp_alpha=0.0, class_weight=None,
criterion='gini', max_depth=None, max_features='auto',
max_leaf_nodes=None, max_samples=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=8,
n_jobs=None, oob_score=False, random_state=None,
verbose=0, warm_start=False)
training took 0.044998s!
precision: 94.83%, recall: 94.02%
accuracy: 93.09%
classification_report:
precision recall f1-score support
0 0.90 0.92 0.91 71
1 0.95 0.94 0.94 117
accuracy 0.93 188
macro avg 0.93 0.93 0.93 188
weighted avg 0.93 0.93 0.93 188
confusion_matrix:
[[ 65 6]
[ 7 110]]
elaspe: 1.873387
******************* DT(决策树) ********************
DecisionTreeClassifier(ccp_alpha=0.0, class_weight=None, criterion='gini',
max_depth=None, max_features=None, max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, presort='deprecated',
random_state=None, splitter='best')
training took 0.014005s!
precision: 93.16%, recall: 93.16%
accuracy: 91.49%
classification_report:
precision recall f1-score support
0 0.89 0.89 0.89 71
1 0.93 0.93 0.93 117
accuracy 0.91 188
macro avg 0.91 0.91 0.91 188
weighted avg 0.91 0.91 0.91 188
confusion_matrix:
[[ 63 8]
[ 8 109]]
elaspe: 0.448771
******************* SVM(支持向量机) ********************
SVC(C=1.0, break_ties=False, cache_size=200, class_weight=None, coef0=0.0,
decision_function_shape='ovr', degree=3, gamma='scale', kernel='rbf',
max_iter=-1, probability=True, random_state=None, shrinking=True, tol=0.001,
verbose=False)
training took 0.028140s!
precision: 90.48%, recall: 97.44%
accuracy: 92.02%
classification_report:
precision recall f1-score support
0 0.95 0.83 0.89 71
1 0.90 0.97 0.94 117
accuracy 0.92 188
macro avg 0.93 0.90 0.91 188
weighted avg 0.92 0.92 0.92 188
confusion_matrix:
[[ 59 12]
[ 3 114]]
elaspe: 1.027975
******************* GBDT(梯度提升决策树) ********************
GradientBoostingClassifier(ccp_alpha=0.0, criterion='friedman_mse', init=None,
learning_rate=0.1, loss='deviance', max_depth=3,
max_features=None, max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=200,
n_iter_no_change=None, presort='deprecated',
random_state=None, subsample=1.0, tol=0.0001,
validation_fraction=0.1, verbose=0,
warm_start=False)
training took 0.996242s!
precision: 94.07%, recall: 94.87%
accuracy: 93.09%
classification_report:
precision recall f1-score support
0 0.91 0.90 0.91 71
1 0.94 0.95 0.94 117
accuracy 0.93 188
macro avg 0.93 0.93 0.93 188
weighted avg 0.93 0.93 0.93 188
confusion_matrix:
[[ 64 7]
[ 6 111]]
elaspe: 39.072309
******************* Adaboost ********************
AdaBoostClassifier(algorithm='SAMME',
base_estimator=DecisionTreeClassifier(ccp_alpha=0.0,
class_weight=None,
criterion='gini',
max_depth=None,
max_features=None,
max_leaf_nodes=None,
min_impurity_decrease=0.0,
min_impurity_split=None,
min_samples_leaf=1,
min_samples_split=2,
min_weight_fraction_leaf=0.0,
presort='deprecated',
random_state=None,
splitter='best'),
learning_rate=0.4, n_estimators=7, random_state=None)
training took 0.025010s!
precision: 93.22%, recall: 94.02%
accuracy: 92.02%
classification_report:
precision recall f1-score support
0 0.90 0.89 0.89 71
1 0.93 0.94 0.94 117
accuracy 0.92 188
macro avg 0.92 0.91 0.91 188
weighted avg 0.92 0.92 0.92 188
confusion_matrix:
[[ 63 8]
[ 7 110]]
elaspe: 0.960197
******************* Bagging ********************
BaggingClassifier(base_estimator=DecisionTreeClassifier(ccp_alpha=0.0,
class_weight=None,
criterion='gini',
max_depth=None,
max_features=None,
max_leaf_nodes=None,
min_impurity_decrease=0.0,
min_impurity_split=None,
min_samples_leaf=1,
min_samples_split=2,
min_weight_fraction_leaf=0.0,
presort='deprecated',
random_state=None,
splitter='best'),
bootstrap=True, bootstrap_features=False, max_features=1.0,
max_samples=1.0, n_estimators=10, n_jobs=None,
oob_score=False, random_state=None, verbose=0,
warm_start=False)
training took 0.106950s!
precision: 94.02%, recall: 94.02%
accuracy: 92.55%
classification_report:
precision recall f1-score support
0 0.90 0.90 0.90 71
1 0.94 0.94 0.94 117
accuracy 0.93 188
macro avg 0.92 0.92 0.92 188
weighted avg 0.93 0.93 0.93 188
confusion_matrix:
[[ 64 7]
[ 7 110]]
elaspe: 4.000736
当使用默认参数时, GBDT(梯度提升决策树)的准确率和召回率最高,同时耗费的时间也最长;相对的MNB(多项式分布朴素贝叶斯)耗费的时间最短。
5.参数调优
各个分类模型的默认参数
KNN Classifier
KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski',
metric_params=None, n_jobs=None, n_neighbors=5, p=2,
weights='uniform')
userscript.html?id=1cfc3476-717c-41b6-b4e7-1a24541c7949:24
scikit-learn K近邻法类库使用小结 - 刘建平Pinard - 博客园
Logistic Regression Classifier
LogisticRegression(C=1.0, class_weight=None, dual=False, fit_intercept=True,
intercept_scaling=1, l1_ratio=None, max_iter=100,
multi_class='auto', n_jobs=None, penalty='l2',
random_state=None, solver='lbfgs', tol=0.0001, verbose=0,
warm_start=False)
Random Forest Classifier
RandomForestClassifier(bootstrap=True, ccp_alpha=0.0, class_weight=None,
criterion='gini', max_depth=None, max_features='auto',
max_leaf_nodes=None, max_samples=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=8,
n_jobs=None, oob_score=False, random_state=None,
verbose=0, warm_start=False)
Decision Tree Classifier
max_depth(树的深度)
max_leaf_nodes(叶子结点的数目)
max_features(最大特征数目)
min_samples_leaf(叶子结点的最小样本数)
min_samples_split(中间结点的最小样本树)
min_weight_fraction_leaf(叶子节点的样本权重占总权重的比例)
min_impurity_split(最小不纯净度)也可以调整
DecisionTreeClassifier(ccp_alpha=0.0, class_weight=None, criterion='gini',
max_depth=None, max_features=None, max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, presort='deprecated',
random_state=None, splitter='best')
[sklearn决策树之剪枝参数_数据结构与算法_The Zen of Data Analysis-CSDN博客](https://blog.csdn.net/gracejpw/article/details/102239574)
SKlearn中分类决策树的重要参数详解—简书
GBDT(Gradient Boosting Decision Tree) Classifier
GradientBoostingClassifier(ccp_alpha=0.0, criterion='friedman_mse', init=None,
learning_rate=0.1, loss='deviance', max_depth=3,
max_features=None, max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=200,
n_iter_no_change=None, presort='deprecated',
random_state=None, subsample=1.0, tol=0.0001,
validation_fraction=0.1, verbose=0,
warm_start=False)
AdaBoost
AdaBoostClassifier(algorithm='SAMME',
base_estimator=DecisionTreeClassifier(ccp_alpha=0.0,
class_weight=None,
criterion='gini',
max_depth=None,
max_features=None,
max_leaf_nodes=None,
min_impurity_decrease=0.0,
min_impurity_split=None,
min_samples_leaf=1,
min_samples_split=2,
min_weight_fraction_leaf=0.0,
presort='deprecated',
random_state=None,
splitter='best'),
learning_rate=0.4, n_estimators=7, random_state=None)
Bagging
BaggingClassifier(base_estimator=DecisionTreeClassifier(ccp_alpha=0.0,
class_weight=None,
criterion='gini',
max_depth=None,
max_features=None,
max_leaf_nodes=None,
min_impurity_decrease=0.0,
min_impurity_split=None,
min_samples_leaf=1,
min_samples_split=2,
min_weight_fraction_leaf=0.0,
presort='deprecated',
random_state=None,
splitter='best'),
bootstrap=True, bootstrap_features=False, max_features=1.0,
max_samples=1.0, n_estimators=10, n_jobs=None,
oob_score=False, random_state=None, verbose=0,
warm_start=False)
SVM
SVC(C=1.0, break_ties=False, cache_size=200, class_weight=None, coef0=0.0,
decision_function_shape='ovr', degree=3, gamma='scale', kernel='rbf',
max_iter=-1, probability=True, random_state=None, shrinking=True, tol=0.001,
verbose=False)
机器学习之SVM调参实例—|旧市拾荒|—博客园
机器学习各种算法怎么调参?—知乎
GBDT、XGBoost、LightGBM 的使用及参数调优 - 简书
[Python嗯~机器学习]—sklearn中对于梯度提升树GBDT和随机森林RF的参数调优_人工智能_kepengs的博客-CSDN博客
def grid_search(model,param_grid,train_x,train_y,cv=5):
grid_search = GridSearchCV(model, param_grid=param_grid, n_jobs = -1, verbose=1) # cv:交叉验证参数,默认是None, 使用三折交叉验证,指定 fold数量, default = 3
grid_search.fit(train_x, train_y)
best_parameters = grid_search.best_estimator_.get_params()
# for para, val in list(best_parameters.items()):
# print(para, val)
print('最优参数:',best_parameters)
return grid_search.best_estimator_
# 调整参数的字典
common_classifiers = ['KNN(最近邻)', 'LR(Logistic回归)', 'DT(决策树)', 'SVM(支持向量机)' ]
ensem_classifiers = ['RF(随机森林)','GBDT(梯度提升决策树)','Adaboost']
basic_classifiers = {
'KNN(最近邻)':KNeighborsClassifier(),
'LR(Logistic回归)':LogisticRegression(penalty='l2'),
'DT(决策树)': DecisionTreeClassifier() ,
'SVM(支持向量机)': SVC(kernel='rbf', probability=True),
'GBDT(梯度提升决策树)': GradientBoostingClassifier(n_estimators=200),
'RF(随机森林)': RandomForestClassifier(n_estimators=8) ,
'Adaboost': AdaBoostClassifier(DecisionTreeClassifier(),algorithm="SAMME", n_estimators=7, learning_rate=0.4)
}
grid_params = {
'KNN(最近邻)':[
{'weights':['uniform'],'n_neighbors':np.arange(4,8,1)},
{'weights':['distance'],'n_neighbors':np.arange(4,8,1)},
],
'LR(Logistic回归)':[
{'C':[0.01,0.1,1.0,10.0,100.0],'penalty':['l1']},
{'C':[0.01,0.1,1.0,10.0,100.0],'penalty':['l2'],'solver':['liblinear','newton-cg','sag','lbfgs']},
],
'DT(决策树)':[
{'min_samples_split':np.arange(1,15,1),'min_samples_leaf':np.arange(1,15,1),'splitter':['random']},
{'min_samples_split':np.arange(1,15,1),'min_samples_leaf':np.arange(1,15,1),'splitter':['best']},
],
'SVM(支持向量机)':[
{'C': [1e-1, 1, 10, 100, 1000], 'kernel': ['linear']},
{'C': [1e-1, 1, 10, 100, 1000], 'gamma': [0.001, 0.0001], 'kernel': ['rbf']},
]
}
ensem_params = {
'GBDT(梯度提升决策树)':{'n_estimators':np.arange(20,500,50),'max_depth':np.arange(3,14,2), 'min_samples_split':np.arange(2,10,2)},#'min_samples_split':list(range(800,1900,200)), 'min_samples_leaf':list(range(60,101,10))
'RF(随机森林)':{'n_estimators':np.arange(10,71,10),'max_depth':np.arange(3,14,2), 'min_samples_split':np.arange(80,150,20), 'min_samples_leaf':np.arange(10,60,10)},
'Adaboost':{'n_estimators':np.arange(1,11,1),'learning_rate':np.arange(0.1,1,0.1)}
}
from sklearn.metrics import roc_curve, auc, roc_auc_score
for classifier in common_classifiers:
print('******************* %s ********************' % classifier)
start_time = time.time()
model = basic_classifiers[classifier]
clf = grid_search(model,grid_params[classifier],x_train,y_train,cv=5)
print('training took %fs!' % (time.time() - start_time))
print(clf)
clf.fit(x_train,y_train)
predict = clf.predict(x_test)
score = metrics.precision_score(y_test, predict)
recall = metrics.recall_score(y_test, predict)
print('precision: %.2f%%, recall: %.2f%%' % (100 * score, 100 * recall))
accuracy = metrics.accuracy_score(y_test, predict)
print('accuracy: %.2f%%' % (100 * accuracy))
c_matrix = confusion_matrix(
y_test, # array, Gound true (correct) target values
predict, # array, Estimated targets as returned by a classifier
labels=[0,1], # array, List of labels to index the matrix.
sample_weight=None # array-like of shape = [n_samples], Optional sample weights
)
print('\nclassification_report:')
print(classification_report( y_test,predict,labels=[0,1]))
print('\nconfusion_matrix:')
print(c_matrix)
print()
curve1 = plot_roc_curve(clf, x_train, y_train, alpha=0.8,name=classifier)
curve1.figure_.suptitle("乳腺癌 ROC")
plt.show()
******************* KNN(最近邻) ********************
Fitting 5 folds for each of 8 candidates, totalling 40 fits
[Parallel(n_jobs=-1)]: Using backend LokyBackend with 4 concurrent workers.
[Parallel(n_jobs=-1)]: Done 40 out of 40 | elapsed: 2.7s finished
最优参数: {'algorithm': 'auto', 'leaf_size': 30, 'metric': 'minkowski', 'metric_params': None, 'n_jobs': None, 'n_neighbors': 6, 'p': 2, 'weights': 'uniform'}
training took 2.806710s!
KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski',
metric_params=None, n_jobs=None, n_neighbors=6, p=2,
weights='uniform')
precision: 94.92%, recall: 97.39%
accuracy: 95.21%
classification_report:
precision recall f1-score support
0 0.96 0.92 0.94 73
1 0.95 0.97 0.96 115
accuracy 0.95 188
macro avg 0.95 0.95 0.95 188
weighted avg 0.95 0.95 0.95 188
confusion_matrix:
[[ 67 6]
[ 3 112]]
******************* LR(Logistic回归) ********************
Fitting 5 folds for each of 25 candidates, totalling 125 fits
[Parallel(n_jobs=-1)]: Using backend LokyBackend with 4 concurrent workers.
[Parallel(n_jobs=-1)]: Done 125 out of 125 | elapsed: 3.8s finished
最优参数: {'C': 100.0, 'class_weight': None, 'dual': False, 'fit_intercept': True, 'intercept_scaling': 1, 'l1_ratio': None, 'max_iter': 100, 'multi_class': 'auto', 'n_jobs': None, 'penalty': 'l2', 'random_state': None, 'solver': 'liblinear', 'tol': 0.0001, 'verbose': 0, 'warm_start': False}
training took 3.939845s!
LogisticRegression(C=100.0, class_weight=None, dual=False, fit_intercept=True,
intercept_scaling=1, l1_ratio=None, max_iter=100,
multi_class='auto', n_jobs=None, penalty='l2',
random_state=None, solver='liblinear', tol=0.0001, verbose=0,
warm_start=False)
precision: 95.83%, recall: 100.00%
accuracy: 97.34%
classification_report:
precision recall f1-score support
0 1.00 0.93 0.96 73
1 0.96 1.00 0.98 115
accuracy 0.97 188
macro avg 0.98 0.97 0.97 188
weighted avg 0.97 0.97 0.97 188
confusion_matrix:
[[ 68 5]
[ 0 115]]
******************* DT(决策树) ********************
Fitting 5 folds for each of 392 candidates, totalling 1960 fits
[Parallel(n_jobs=-1)]: Using backend LokyBackend with 4 concurrent workers.
[Parallel(n_jobs=-1)]: Done 312 tasks | elapsed: 0.9s
[Parallel(n_jobs=-1)]: Done 1960 out of 1960 | elapsed: 4.6s finished
最优参数: {'ccp_alpha': 0.0, 'class_weight': None, 'criterion': 'gini', 'max_depth': None, 'max_features': None, 'max_leaf_nodes': None, 'min_impurity_decrease': 0.0, 'min_impurity_split': None, 'min_samples_leaf': 1, 'min_samples_split': 4, 'min_weight_fraction_leaf': 0.0, 'presort': 'deprecated', 'random_state': None, 'splitter': 'random'}
training took 4.746069s!
DecisionTreeClassifier(ccp_alpha=0.0, class_weight=None, criterion='gini',
max_depth=None, max_features=None, max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=4,
min_weight_fraction_leaf=0.0, presort='deprecated',
random_state=None, splitter='random')
precision: 95.54%, recall: 93.04%
accuracy: 93.09%
classification_report:
precision recall f1-score support
0 0.89 0.93 0.91 73
1 0.96 0.93 0.94 115
accuracy 0.93 188
macro avg 0.93 0.93 0.93 188
weighted avg 0.93 0.93 0.93 188
confusion_matrix:
[[ 68 5]
[ 8 107]]
******************* SVM(支持向量机) ********************
Fitting 5 folds for each of 15 candidates, totalling 75 fits
[Parallel(n_jobs=-1)]: Using backend LokyBackend with 4 concurrent workers.
[Parallel(n_jobs=-1)]: Done 42 tasks | elapsed: 7.4min
[Parallel(n_jobs=-1)]: Done 75 out of 75 | elapsed: 10.5min finished
最优参数: {'C': 10, 'break_ties': False, 'cache_size': 200, 'class_weight': None, 'coef0': 0.0, 'decision_function_shape': 'ovr', 'degree': 3, 'gamma': 'scale', 'kernel': 'linear', 'max_iter': -1, 'probability': True, 'random_state': None, 'shrinking': True, 'tol': 0.001, 'verbose': False}
training took 698.401171s!
SVC(C=10, break_ties=False, cache_size=200, class_weight=None, coef0=0.0,
decision_function_shape='ovr', degree=3, gamma='scale', kernel='linear',
max_iter=-1, probability=True, random_state=None, shrinking=True, tol=0.001,
verbose=False)
precision: 96.58%, recall: 98.26%
accuracy: 96.81%
classification_report:
precision recall f1-score support
0 0.97 0.95 0.96 73
1 0.97 0.98 0.97 115
accuracy 0.97 188
macro avg 0.97 0.96 0.97 188
weighted avg 0.97 0.97 0.97 188
confusion_matrix:
[[ 69 4]
[ 2 113]]
集成学习调参
for classifier in ensem_classifiers:
print('******************* %s ********************' % classifier)
start_time = time.time()
model = basic_classifiers[classifier]
clf = grid_search(model,ensem_params[classifier],x_train,y_train,cv=5)
print('training took %fs!' % (time.time() - start_time))
print(clf)
clf.fit(x_train,y_train)
predict = clf.predict(x_test)
score = metrics.precision_score(y_test, predict)
recall = metrics.recall_score(y_test, predict)
print('precision: %.2f%%, recall: %.2f%%' % (100 * score, 100 * recall))
accuracy = metrics.accuracy_score(y_test, predict)
print('accuracy: %.2f%%' % (100 * accuracy))
c_matrix = confusion_matrix(
y_test, # array, Gound true (correct) target values
predict, # array, Estimated targets as returned by a classifier
labels=[0,1], # array, List of labels to index the matrix.
sample_weight=None # array-like of shape = [n_samples], Optional sample weights
)
print('\nclassification_report:')
print(classification_report( y_test,predict,labels=[0,1]))
print('\nconfusion_matrix:')
print(c_matrix)
print()
curve1 = plot_roc_curve(clf, x_train, y_train, alpha=0.8,name=classifier)
curve1.figure_.suptitle("乳腺癌 ROC")
plt.show()
******************* RF(随机森林) ********************
Fitting 5 folds for each of 840 candidates, totalling 4200 fits
[Parallel(n_jobs=-1)]: Using backend LokyBackend with 4 concurrent workers.
[Parallel(n_jobs=-1)]: Done 42 tasks | elapsed: 4.7s
[Parallel(n_jobs=-1)]: Done 192 tasks | elapsed: 12.4s
[Parallel(n_jobs=-1)]: Done 442 tasks | elapsed: 27.4s
[Parallel(n_jobs=-1)]: Done 792 tasks | elapsed: 55.1s
[Parallel(n_jobs=-1)]: Done 1242 tasks | elapsed: 1.5min
[Parallel(n_jobs=-1)]: Done 1792 tasks | elapsed: 2.2min
[Parallel(n_jobs=-1)]: Done 2442 tasks | elapsed: 3.0min
[Parallel(n_jobs=-1)]: Done 3192 tasks | elapsed: 3.8min
[Parallel(n_jobs=-1)]: Done 4042 tasks | elapsed: 4.8min
[Parallel(n_jobs=-1)]: Done 4200 out of 4200 | elapsed: 5.0min finished
最优参数: {'bootstrap': True, 'ccp_alpha': 0.0, 'class_weight': None, 'criterion': 'gini', 'max_depth': 3, 'max_features': 'auto', 'max_leaf_nodes': None, 'max_samples': None, 'min_impurity_decrease': 0.0, 'min_impurity_split': None, 'min_samples_leaf': 10, 'min_samples_split': 120, 'min_weight_fraction_leaf': 0.0, 'n_estimators': 30, 'n_jobs': None, 'oob_score': False, 'random_state': None, 'verbose': 0, 'warm_start': False}
training took 301.064679s!
RandomForestClassifier(bootstrap=True, ccp_alpha=0.0, class_weight=None,
criterion='gini', max_depth=3, max_features='auto',
max_leaf_nodes=None, max_samples=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=10, min_samples_split=120,
min_weight_fraction_leaf=0.0, n_estimators=30,
n_jobs=None, oob_score=False, random_state=None,
verbose=0, warm_start=False)
precision: 94.17%, recall: 94.17%
accuracy: 92.55%
classification_report:
precision recall f1-score support
0 0.90 0.90 0.90 68
1 0.94 0.94 0.94 120
accuracy 0.93 188
macro avg 0.92 0.92 0.92 188
weighted avg 0.93 0.93 0.93 188
confusion_matrix:
[[ 61 7]
[ 7 113]]
******************* GBDT(梯度提升决策树) ********************
Fitting 5 folds for each of 240 candidates, totalling 1200 fits
[Parallel(n_jobs=-1)]: Using backend LokyBackend with 4 concurrent workers.
[Parallel(n_jobs=-1)]: Done 42 tasks | elapsed: 11.9s
[Parallel(n_jobs=-1)]: Done 192 tasks | elapsed: 1.0min
[Parallel(n_jobs=-1)]: Done 442 tasks | elapsed: 2.1min
[Parallel(n_jobs=-1)]: Done 792 tasks | elapsed: 3.8min
[Parallel(n_jobs=-1)]: Done 1200 out of 1200 | elapsed: 5.2min finished
最优参数: {'ccp_alpha': 0.0, 'criterion': 'friedman_mse', 'init': None, 'learning_rate': 0.1, 'loss': 'deviance', 'max_depth': 3, 'max_features': None, 'max_leaf_nodes': None, 'min_impurity_decrease': 0.0, 'min_impurity_split': None, 'min_samples_leaf': 1, 'min_samples_split': 2, 'min_weight_fraction_leaf': 0.0, 'n_estimators': 420, 'n_iter_no_change': None, 'presort': 'deprecated', 'random_state': None, 'subsample': 1.0, 'tol': 0.0001, 'validation_fraction': 0.1, 'verbose': 0, 'warm_start': False}
training took 314.633900s!
GradientBoostingClassifier(ccp_alpha=0.0, criterion='friedman_mse', init=None,
learning_rate=0.1, loss='deviance', max_depth=3,
max_features=None, max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=420,
n_iter_no_change=None, presort='deprecated',
random_state=None, subsample=1.0, tol=0.0001,
validation_fraction=0.1, verbose=0,
warm_start=False)
precision: 96.75%, recall: 99.17%
accuracy: 97.34%
classification_report:
precision recall f1-score support
0 0.98 0.94 0.96 68
1 0.97 0.99 0.98 120
accuracy 0.97 188
macro avg 0.98 0.97 0.97 188
weighted avg 0.97 0.97 0.97 188
confusion_matrix:
[[ 64 4]
[ 1 119]]
******************* Adaboost ********************
Fitting 5 folds for each of 90 candidates, totalling 450 fits
[Parallel(n_jobs=-1)]: Using backend LokyBackend with 4 concurrent workers.
[Parallel(n_jobs=-1)]: Done 280 tasks | elapsed: 1.4s
[Parallel(n_jobs=-1)]: Done 450 out of 450 | elapsed: 2.0s finished
最优参数: {'algorithm': 'SAMME', 'base_estimator__ccp_alpha': 0.0, 'base_estimator__class_weight': None, 'base_estimator__criterion': 'gini', 'base_estimator__max_depth': None, 'base_estimator__max_features': None, 'base_estimator__max_leaf_nodes': None, 'base_estimator__min_impurity_decrease': 0.0, 'base_estimator__min_impurity_split': None, 'base_estimator__min_samples_leaf': 1, 'base_estimator__min_samples_split': 2, 'base_estimator__min_weight_fraction_leaf': 0.0, 'base_estimator__presort': 'deprecated', 'base_estimator__random_state': None, 'base_estimator__splitter': 'best', 'base_estimator': DecisionTreeClassifier(ccp_alpha=0.0, class_weight=None, criterion='gini',
max_depth=None, max_features=None, max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, presort='deprecated',
random_state=None, splitter='best'), 'learning_rate': 0.7000000000000001, 'n_estimators': 3, 'random_state': None}
training took 2.143543s!
AdaBoostClassifier(algorithm='SAMME',
base_estimator=DecisionTreeClassifier(ccp_alpha=0.0,
class_weight=None,
criterion='gini',
max_depth=None,
max_features=None,
max_leaf_nodes=None,
min_impurity_decrease=0.0,
min_impurity_split=None,
min_samples_leaf=1,
min_samples_split=2,
min_weight_fraction_leaf=0.0,
presort='deprecated',
random_state=None,
splitter='best'),
learning_rate=0.7000000000000001, n_estimators=3,
random_state=None)
precision: 95.04%, recall: 95.83%
accuracy: 94.15%
classification_report:
precision recall f1-score support
0 0.93 0.91 0.92 68
1 0.95 0.96 0.95 120
accuracy 0.94 188
macro avg 0.94 0.94 0.94 188
weighted avg 0.94 0.94 0.94 188
confusion_matrix:
[[ 62 6]
[ 5 115]]
经过对比发现,通过网格寻优对参数进行调参后,模型的准确率有所上升。