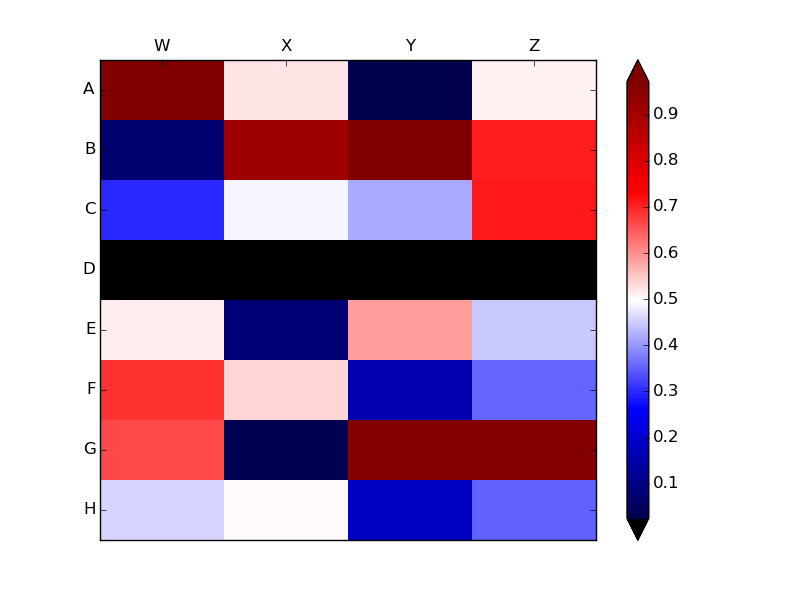
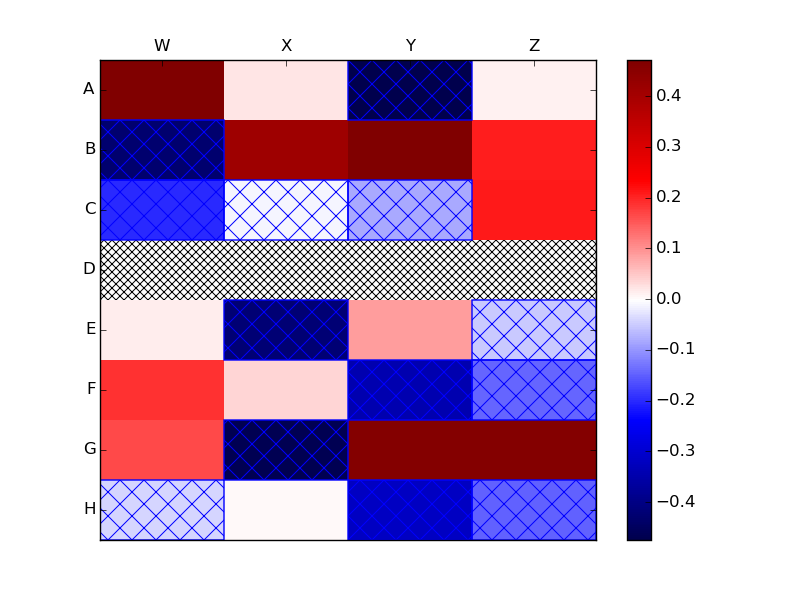
python绘制色块图_Python:从matplotlib热图及其图例中保留Numpy NaN值
nans干扰pcolor确定数据中包含的值的范围
In [72]: data.min(), data.max()
Out[72]: (nan, nan)
您可以通过使用np.nanmin和np.nanmax自行声明值范围来查找数据中的最小和最大非NaN值来解决此问题:
heatmap = ax.pcolor(data, cmap=plt.cm.seismic,
vmin=np.nanmin(data), vmax=np.nanmax(data))
以来
In [73]: np.nanmin(data), np.nanmax(data)
Out[73]: (0.025462800000000001, 0.97094435999999995)
import numpy as np
import matplotlib.pyplot as plt
column_labels = list('ABCDEFGH')
row_labels = list('WXYZ')
fig, ax = plt.subplots()
data = np.array([[ 0.96753494, 0.52349944, 0.0254628 , 0.5104103 ],
[ 0.07320069, 0.91278731, 0.97094436, 0.70533351],
[ 0.30162006, 0.49068337, 0.41837729, 0.71139215],
[ 0.19786101, 0.15882713, 0.59028841, 0.06242765],
[ 0.51505872, 0.07798389, 0.58790067, 0.44782683],
[ 0.68975694, 0.53535385, 0.15696023, 0.35641951],
[ 0.66481995, 0.03576846, 0.9623601 , 0.96006395],
[ 0.45865404, 0.50433582, 0.18182575, 0.35126449],])
data[3,:] = np.nan
heatmap = ax.pcolor(data, cmap=plt.cm.seismic,
vmin=np.nanmin(data), vmax=np.nanmax(data))
heatmap.cmap.set_under('black')
bar = fig.colorbar(heatmap, extend='both')
# put the major ticks at the middle of each cell
ax.set_xticks(np.arange(data.shape[1])+0.5, minor=False)
ax.set_yticks(np.arange(data.shape[0])+0.5, minor=False)
# want a more natural, table-like display
ax.invert_yaxis()
ax.xaxis.tick_top()
ax.set_xticklabels(row_labels, minor=False)
ax.set_yticklabels(column_labels, minor=False)
plt.show()
另一种选择(基于Joe Kington’s
solution)将是绘制
数据为NaN的带有阴影标记的矩形色块.
上面的例子显示了NaN值为的单元格中的pcolor颜色
虽然NaNs是非常负数.相反,如果你通过pcolor a
掩蔽的数组,pcolor使蒙面区域透明.因此,你可以画画
轴背景补丁,ax.patch上的阴影,以显示阴影标记
在蒙面区域.
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
column_labels = list('ABCDEFGH')
row_labels = list('WXYZ')
fig, ax = plt.subplots()
data = np.array([[ 0.96753494, 0.52349944, 0.0254628 , 0.5104103 ],
[ 0.07320069, 0.91278731, 0.97094436, 0.70533351],
[ 0.30162006, 0.49068337, 0.41837729, 0.71139215],
[ 0.19786101, 0.15882713, 0.59028841, 0.06242765],
[ 0.51505872, 0.07798389, 0.58790067, 0.44782683],
[ 0.68975694, 0.53535385, 0.15696023, 0.35641951],
[ 0.66481995, 0.03576846, 0.9623601 , 0.96006395],
[ 0.45865404, 0.50433582, 0.18182575, 0.35126449],])
data[3,:] = np.nan
data = np.ma.masked_invalid(data)
heatmap = ax.pcolor(data, cmap=plt.cm.seismic,
vmin=np.nanmin(data), vmax=np.nanmax(data))
# https://stackoverflow.com/a/16125413/190597 (Joe Kington)
ax.patch.set(hatch='x', edgecolor='black')
fig.colorbar(heatmap)
# put the major ticks at the middle of each cell
ax.set_xticks(np.arange(data.shape[1])+0.5, minor=False)
ax.set_yticks(np.arange(data.shape[0])+0.5, minor=False)
# want a more natural, table-like display
ax.invert_yaxis()
ax.xaxis.tick_top()
ax.set_xticklabels(row_labels, minor=False)
ax.set_yticklabels(column_labels, minor=False)
plt.show()
如果您希望使用多种类型的阴影标记,例如一个用于NaN而另一个用于负值,则可以使用循环添加阴影线矩形:
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
column_labels = list('ABCDEFGH')
row_labels = list('WXYZ')
fig, ax = plt.subplots()
data = np.array([[ 0.96753494, 0.52349944, 0.0254628 , 0.5104103 ],
[ 0.07320069, 0.91278731, 0.97094436, 0.70533351],
[ 0.30162006, 0.49068337, 0.41837729, 0.71139215],
[ 0.19786101, 0.15882713, 0.59028841, 0.06242765],
[ 0.51505872, 0.07798389, 0.58790067, 0.44782683],
[ 0.68975694, 0.53535385, 0.15696023, 0.35641951],
[ 0.66481995, 0.03576846, 0.9623601 , 0.96006395],
[ 0.45865404, 0.50433582, 0.18182575, 0.35126449],])
data -= 0.5
data[3,:] = np.nan
data = np.ma.masked_invalid(data)
heatmap = ax.pcolor(data, cmap=plt.cm.seismic,
vmin=np.nanmin(data), vmax=np.nanmax(data))
# https://stackoverflow.com/a/16125413/190597 (Joe Kington)
ax.patch.set(hatch='x', edgecolor='black')
# draw a hatched rectangle wherever the data is negative
# http://matthiaseisen.com/pp/patterns/p0203/
mask = data < 0
for j, i in np.column_stack(np.where(mask)):
ax.add_patch(
mpatches.Rectangle(
(i, j), # (x,y)
1, # width
1, # height
fill=False,
edgecolor='blue',
snap=False,
hatch='x' # the more slashes, the denser the hash lines
))
fig.colorbar(heatmap)
# put the major ticks at the middle of each cell
ax.set_xticks(np.arange(data.shape[1])+0.5, minor=False)
ax.set_yticks(np.arange(data.shape[0])+0.5, minor=False)
# want a more natural, table-like display
ax.invert_yaxis()
ax.xaxis.tick_top()
ax.set_xticklabels(row_labels, minor=False)
ax.set_yticklabels(column_labels, minor=False)
plt.show()