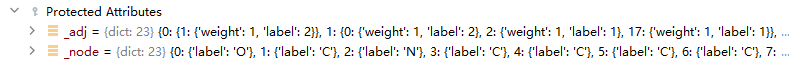分子的建图(smiles字符串、networkx图、dgl图)
目录
-
- 头文件
- smiles字符串
- 转化为networkx中的图
-
- networkx图的绘制
- 加节点属性
- 加边属性
- networkx访问边的属性
- networkx图转化为dgl图
- 直接根据分子图构建dgl图
-
- 构建batch
- 加载和保存
- 部分参考
头文件
import rdkit.Chem as Chem
import rdkit
import networkx as nx
import matplotlib.pyplot as plt
import numpy as np
import dgl
smiles字符串
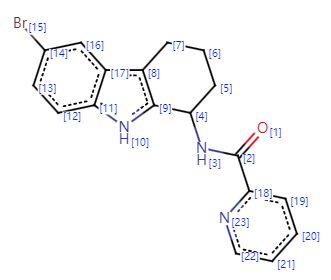
给定一个经过 canonicalize的smiles字符串(原子编号从1开始):
smiles = '[O:1]=[C:2]([NH:3][CH:4]1[CH2:5][CH2:6][CH2:7][c:8]2[c:9]1[nH:10][c:11]1[cH:12][cH:13][c:14]([Br:15])[cH:16][c:17]21)[c:18]1[cH:19][cH:20][cH:21][cH:22][n:23]1'
表示如下分子:(通过https://marvinjs-demo.chemaxon.com/latest/可视化)
转化为networkx中的图
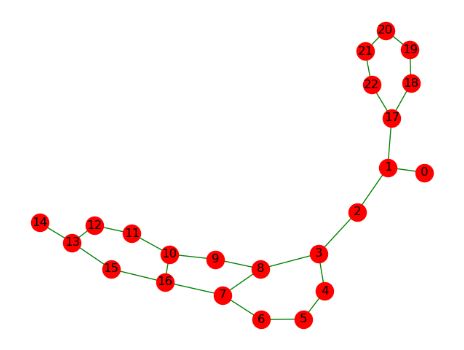
networkx图的绘制
mol = Chem.MolFromSmiles(smiles)
G_networkx = nx.Graph(Chem.rdmolops.GetAdjacencyMatrix(mol))
nx.draw(G_networkx,with_labels=True,node_color="r",edge_color="g")
plt.show()
在这里,通过传入类型为ndarray的邻接矩阵来得到networkx中的无向图Graph

通过matplotlib进行可视化(节点编号从0开始):
加节点属性
for atom in mol.GetAtoms():
G_networkx.nodes[atom.GetIdx()]['label'] = atom.GetSymbol()
加边属性
BOND_TYPES = [None, Chem.rdchem.BondType.SINGLE, Chem.rdchem.BondType.DOUBLE, \
Chem.rdchem.BondType.TRIPLE, Chem.rdchem.BondType.AROMATIC]
for bond in mol.GetBonds():
a1 = bond.GetBeginAtom().GetIdx()
a2 = bond.GetEndAtom().GetIdx()
btype = BOND_TYPES.index(bond.GetBondType())
G_networkx[a1][a2]['label'] = btype
networkx访问边的属性
edge_u = []
edge_v = []
attr_uv = []
for (u,v,d) in G_networkx.edges(data=True):
edge_u.append(u)
edge_v.append(v)
attr_uv.append(d)
节点属性类似
访问具体某条边,需要确定边的id
g.edata(g.edge_id(0,7))
networkx图转化为dgl图
dgl可直接根据networkx图构建图。但对于大规模图来说,直接转化效率并不高,更推荐先将networkx图转化为节点向量的元组,再依此构建dgl图。
G_networkx = G_networkx.to_directed()
G_dgl = dgl.from_networkx(G_networkx) # 仅保留图结构
G_dgl = dgl.from_networkx(G_networkx, node_attrs=[], edge_attrs=['label']) #把边的属性也移过去了
dgl图始终是有向的,先将无向图G_networkx转为有向图(如果不转为有向图,在转化为dgl图后还是会变为有向图,但是边的属性没有补齐)。
注意,在dgl中节点和边的属性应该是要能转换为tensor类型的(如int, numpy.ndarray, list等),在上述代码中,由于节点的label属性类型为str,所以会报错。可将上面代码中的节点属性改为原子对应的one-hot编码。
dgl图转化回networkx图:
G_networkx2 = G_dgl.to_networkx()
直接根据分子图构建dgl图
假设节点和边上的属性都为int值
import rdkit.Chem as Chem
import rdkit
import networkx as nx
import matplotlib.pyplot as plt
import numpy as np
import dgl
import torch
smiles = '[O:1]=[C:2]([NH:3][CH:4]1[CH2:5][CH2:6][CH2:7][c:8]2[c:9]1[nH:10][c:11]1[cH:12][cH:13][c:14]([Br:15])[cH:16][c:17]21)[c:18]1[cH:19][cH:20][cH:21][cH:22][n:23]1'
mol = Chem.MolFromSmiles(smiles)
BOND_TYPES = [None, Chem.rdchem.BondType.SINGLE, Chem.rdchem.BondType.DOUBLE, \
Chem.rdchem.BondType.TRIPLE, Chem.rdchem.BondType.AROMATIC]
ATOM_LIST = ['C', 'N', 'O', 'S', 'F', 'Si', 'P', 'Cl', 'Br', 'Mg', 'Na', 'Ca', 'Fe', \
'As', 'Al', 'I', 'B', 'V', 'K', 'Tl', 'Yb', 'Sb', 'Sn', 'Ag', 'Pd', 'Co', 'Se', 'Ti', \
'Zn', 'H', 'Li', 'Ge', 'Cu', 'Au', 'Ni', 'Cd', 'In', 'Mn', 'Zr', 'Cr', 'Pt', 'Hg', 'Pb', \
'W', 'Ru', 'Nb', 'Re', 'Te', 'Rh', 'Ta', 'Tc', 'Ba', 'Bi', 'Hf', 'Mo', 'U', 'Sm', 'Os', 'Ir', \
'Ce','Gd','Ga','Cs', '*', 'unk']
u = []
v = []
node_attr = {'label':[]}
edge_attr = {'label':[]}
for atom in mol.GetAtoms():
node_attr['label'].append(ATOM_LIST.index(atom.GetSymbol()))
for bond in mol.GetBonds():
a1 = bond.GetBeginAtom().GetIdx()
a2 = bond.GetEndAtom().GetIdx()
btype = BOND_TYPES.index(bond.GetBondType())
u.extend([a1])
v.extend([a2])
edge_attr['label'].extend([btype])
G = dgl.graph((u, v), num_nodes=mol.GetNumAtoms())
G.ndata['label'] = torch.tensor(node_attr['label'])
G.edata['label'] = torch.tensor(edge_attr['label'])
U_G = dgl.add_reverse_edges(G,copy_edata=True) #构建无向图 即双向边 保留边数据,复制到反向边
G:
Graph(num_nodes=23, num_edges=26,
ndata_schemes={'label': Scheme(shape=(), dtype=torch.int64)}
edata_schemes={'label': Scheme(shape=(), dtype=torch.int64)})
U_G:
Graph(num_nodes=23, num_edges=52,
ndata_schemes={'label': Scheme(shape=(), dtype=torch.int64)}
edata_schemes={'label': Scheme(shape=(), dtype=torch.int64), '_ID': Scheme(shape=(), dtype=torch.int64)})
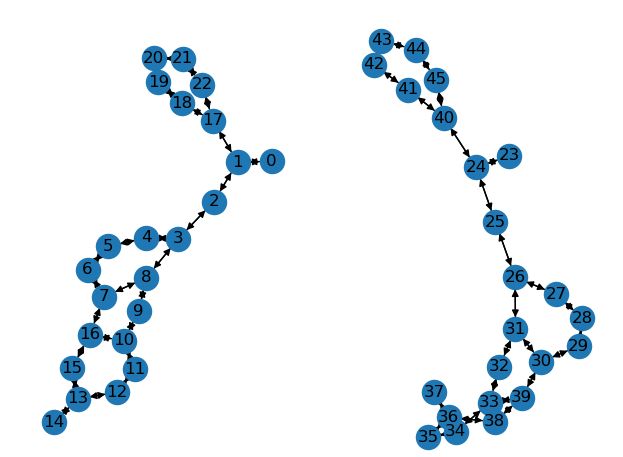
构建batch
graphs = [U_G, U_G]
graph = dgl.batch(graphs)
nx_G = graph.to_networkx()
nx.draw(nx_G, with_labels=True)
plt.show()
在深度学习训练时,需要对数据进行打包。dgl中有个很方便的函数dgl.batch,将多个小图打包成大图,大图的序号重新编号。
加载和保存
dgl.save_graphs('graph.dgl', graph)
(load_g,),_ = dgl.load_graphs('graph.dgl')
print(load_g)
部分参考
networkx是什么
https://docs.dgl.ai/guide/graph-external.html
基于DGL库图神经网络教程(1)——基本的建图操作
DGL的Blitz ---- Blitz的如何用DGL建图 (v9.0 版DGL)
DGL的图数据结构的创建、图的特征、dgl.batch及一些理解