AI制药 - AlphaFold DB PDB 数据集的多维度分析与整理 (2)
欢迎关注我的CSDN:https://spike.blog.csdn.net/
本文地址:https://blog.csdn.net/caroline_wendy/article/details/130118339

数据集:AlphaFold Protein Structure Database
下载地址:https://alphafold.ebi.ac.uk/download
AlphaFold DB 数据量,由最初214M,清洗高置信度为61M,再聚类为5M。
1. AlphaFold DB 官网
数据包含2个部分:
- Compressed prediction files
- UniProt
1.1 Compressed prediction files
The AlphaFold DB website currently provides bulk downloads for the 48 organisms listed below, as well as the majority of Swiss-Prot.
- AlphaFold DB 网站目前提供下列 48 种生物以及大部分 Swiss-Prot 的批量下载。
- Swiss Institute of Bioinformatics,瑞士生物信息学研究所,Prot -> Protein
UniProtKB/Swiss-Prot is the expertly curated component of UniProtKB (produced by the UniProt consortium). It contains hundreds of thousands of protein descriptions, including function, domain structure, subcellular location, post-translational modifications and functionally characterized variants.
UniProtKB/Swiss-Prot 是 UniProtKB(由 UniProt 联盟制作)的专业策划组件,包含数十万种蛋白质描述,包括功能、域结构、亚细胞定位、翻译后修饰和功能特征变异。
结构包括 PDB 和 mmCIF 两种类型,数据集包括:
- Compressed prediction files for model organism proteomes,模型生物蛋白质组 的压缩预测文件
- Compressed prediction files for global health proteomes,全球健康蛋白质组 的压缩预测文件
- Compressed prediction files for Swiss-Prot,Swiss-Prot 的压缩预测文件
- MANE Select dataset,MANE 选择数据集,MANE,Matched Annotation (匹配标注) from NCBI and EMBL-EBI
NCBI,The National Center for Biotechnology Information,国家生物技术信息中心
EMBL-EBI,European Molecular Biology Laboratory - European Bioinformatics Institute,欧洲分子生物学实验室 - 欧洲生物信息学研究所
目前版本最近是v4,下载地址:https://ftp.ebi.ac.uk/pub/databases/alphafold/
1.2 UniProt
全量数据集:Full dataset download for AlphaFold Database - UniProt (214M),即2.14亿
数据集说明:
- 全量数据共214M个样本,保存为 1015808 tar 文件,共23TB,解压后得到3*214M个gz文件,再次解压后得到214M个cif和418M个json
- 每个样本共包含三个文件(1个cif,两个json)
- model_v3.cif – contains the atomic coordinates for the predicted protein structure, along with some metadata. Useful references for this file format are the ModelCIF and PDBx/mmCIF project sites.
- confidence_v3.json – contains a confidence metric output by AlphaFold called pLDDT. This provides a number for each residue, indicating how confident AlphaFold is in the local surrounding structure. pLDDT ranges from 0 to 100, where 100 is most confident. This is also contained in the CIF file.
- predicted_aligned_error_v3.json – contains a confidence metric output by AlphaFold called PAE. This provides a number for every pair of residues, which is lower when AlphaFold is more confident in the relative position of the two residues. PAE is more suitable than pLDDT for judging confidence in relative domain placements.
1.3 单个结构
You can download a prediction for an individual UniProt accession by visiting the corresponding structure page.
- 您可以通过访问相应的结构页面,下载单个 UniProt 新增的预测。
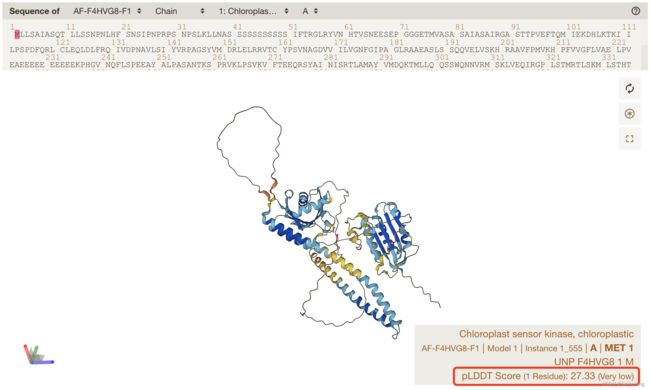
预测的单体PDB结构,例如AF-F4HVG8-F1-model_v4.pdb:
- Chloroplast sensor kinase, chloroplastic:叶绿体传感器激酶, 叶绿体

在AF2 PDB中,Temperature factor (B factor) 温度因子的位置,就是pLDDT的预测值,即
MODEL 1
ATOM 1 N MET A 1 -15.359 18.253 -11.695 1.00 27.33 N
ATOM 2 CA MET A 1 -15.812 17.432 -12.846 1.00 27.33 C
ATOM 3 C MET A 1 -15.487 15.976 -12.539 1.00 27.33 C
ATOM 4 CB MET A 1 -15.064 17.802 -14.142 1.00 27.33 C
ATOM 5 O MET A 1 -14.426 15.760 -11.977 1.00 27.33 O
ATOM 6 CG MET A 1 -15.223 19.246 -14.625 1.00 27.33 C
ATOM 7 SD MET A 1 -14.329 19.504 -16.180 1.00 27.33 S
ATOM 8 CE MET A 1 -14.334 21.313 -16.299 1.00 27.33 C
ATOM 9 N LEU A 2 -16.290 14.967 -12.875 1.00 26.17 N
ATOM 10 CA LEU A 2 -17.714 14.928 -13.250 1.00 26.17 C
ATOM 11 C LEU A 2 -18.221 13.489 -12.989 1.00 26.17 C
ATOM 12 CB LEU A 2 -17.913 15.315 -14.736 1.00 26.17 C
ATOM 13 O LEU A 2 -17.420 12.559 -12.940 1.00 26.17 O
ATOM 14 CG LEU A 2 -18.870 16.504 -14.945 1.00 26.17 C
ATOM 15 CD1 LEU A 2 -18.802 16.990 -16.390 1.00 26.17 C
ATOM 16 CD2 LEU A 2 -20.319 16.141 -14.614 1.00 26.17 C
相对于:
2. 数据清洗
清洗规则:
- 序列长度在 [100, 1000]
- 全局pLDDT在 [90, 100],即
Model Confidence Very High,来自于官网。 - 与 RCSB数据集 去重
遍历一次需要:7775s = 2.15h,样本数量61775031个,7.6G的路径文件,以前2个字母,建立子文件夹。
即数据量由 214M 降至 61M,即由2.14亿降低为6177万
计算PDB的序列长度和pLDDT的值,例如ff文件夹的fffffffb38338e27427f7fef20b3c53f_A.pdb
获取每个原子的pLDDT,参考:BioPython - Bio.PDB.PDBParser:
def get_plddt_from_pdb(self, pdb_path):
p = Bio.PDB.PDBParser()
structure = p.get_structure('', pdb_path)
for a in structure.get_atoms():
print(f"[Info] plddt: {a.get_bfactor()}")
break
获取每个残基的pLDDT,进而计算PDB中残基pLDDT的均值,作为PDB的pLDDT:
def get_plddt_from_pdb(self, pdb_path):
p = Bio.PDB.PDBParser()
structure = p.get_structure('input', pdb_path)
plddt_list = []
for a in structure.get_residues():
b = a.get_unpacked_list()
if len(b) > 0:
plddt_list.append(b[0].get_bfactor())
plddt = np.average(np.array(plddt_list))
plddt = round(plddt, 4)
return plddt
3. 数据聚类
清洗规则:
- 使用mmseq2对6000万进行聚类
- 与v3数据集(plddt>=90的部分)合并,参考 https://ftp.ebi.ac.uk/pub/databases/alphafold/v3/
即由6177万,降低为5040445,即504万,再与v3数据集合并去重,增加至5334594,即533万。
目前版本最近是v4。
4. 信息提取
数据集日志:GitHub deepmind/alphafold - AlphaFold Protein Structure Database
v3版本:2022.7.24,数据路径:gs://public-datasets-deepmind-alphafold

v4版本(最新版本):2022.11.1,数据路径:gs://public-datasets-deepmind-alphafold-v4

整理数据维度,包括:[“pdb”, “plddt”, “release_date”, “seq”, “len”],即
- pdb:pdb的名称
- plddt:置信度
- release_date:pdb数据库时间
- seq:完整序列
- len:长度
具体展示:
id,pdb,plddt,release_date,seq,len
14,9bdf658ff3393c8f5dfeff062ccdef4a_A,90.6749,2022-07-24,MTDSVLDVTADALEQAGAHPHRLVVRQHGQVVGRRRWAPWSPDVPNLAYSCSKTFTSAAVGIAVNRGAFGYDDTLADLWPQACTANTGPVAKSMTVRNALSMSTGHSPEQ,110
11,9bd4cb263bc3b48600fde987daf04fc0_A,93.4367,2022-07-24,MPYADQQAMYDHIDELSQYNAELKSLRSADRVAFRNKYSGQFSMSEIIRRSQIQLKNLHKQRYEVYSDPTLTARQQAVRALMIELNMKKVVDRFYREYREKVGE,104
22,abdda3248f2a38fce73462bd5fd59058_A,91.7146,2022-07-24,DALPTFPDADAFSCIERELGLPLESIFSLISPSPIAAASLGQVYKAQLRYSGQTVAVKVQRPNIEEAVGLDFYLLRNLGFLINKYVDIITSDVVALIDEFARRVYQELNYVQ,112
27,abdda9fde54bdab0eb6bcb447c45f5f0_A,92.8079,2022-07-24,MPAWNVKLTDKIERTSDIISFRFEQPEDLTYLPGQFFFVYIPAEAGGNMMHHFSFSSSPSEPFIEFTTRIRDSPFKKRLNQLEIGTTVEIASVSGQFTLTEEMKKVVFICGGIGITAARSNIKWIIDSHSHSIVDIILLYGNRNYNNIAFKDELEKFTE,159
7,9bd4c9785ede507733f3786f9d20f190_A,94.2027,2022-07-24,MREEVLAAVLAADKELSRDEPEVAANIGKQLEHMGLTTWSISVRRRIRDAIARLEPKHIIQTGSGIGHLSAWILDHFEGSNGLEKFQIVEEGNRFAVILTRLCQRYDSVPTSIKVGAPSLLTSELKAWQISKIGD,135
源码
本源码,也可以复用于其他只有PDB文件,没有PDB信息的数据集中。
#!/usr/bin/env python
# -- coding: utf-8 --
"""
Copyright (c) 2022. All rights reserved.
Created by C. L. Wang on 2023/4/12
"""
import os
import sys
from multiprocessing.pool import Pool
from time import time
import numpy as np
import pandas as pd
from Bio.Data.PDBData import protein_letters_3to1_extended as d3to1_ex
from Bio.PDB import PDBParser
from tqdm import tqdm
p = os.path.dirname(os.path.dirname(os.path.abspath(__file__)))
if p not in sys.path:
sys.path.append(p)
from myutils.project_utils import write_list_to_file, mkdir_if_not_exist, traverse_dir_files_for_large, \
traverse_dir_files, read_file
from root_dir import DATA_DIR
class AfdbProcessor(object):
"""
AlphaFold DB 数据集整理和分析
"""
def __init__(self):
self.out_dir = os.path.join(DATA_DIR, "alphafold_db")
mkdir_if_not_exist(self.out_dir)
# 输入
self.alphafold_dir = "[AlphaFold DB dir]"
self.full_pdb_dir = os.path.join(self.alphafold_dir, "[PDB dir]")
self.pdb_name_path = os.path.join(self.alphafold_dir, "cluster_5M_from_uniprot.txt")
# 输出
full_prefix = "alphafold_db_pdb_all"
self.all_pdb_format = os.path.join(self.out_dir, f"{full_prefix}" + "_{}.txt")
# 读取PDB
paths_list = traverse_dir_files(self.out_dir)
is_traverse = False
for path in paths_list:
base_name = os.path.basename(path)
if full_prefix in base_name:
is_traverse = True
break
if not is_traverse:
self.init_full_paths() # 初始化全部路径
else:
print("[Info] 已经初始化完成PDB全部路径!")
def init_full_paths(self):
print(f"[Info] 初始化路径开始!")
s_time = time()
print(f"[Info] 数据集路径: {self.full_pdb_dir}")
paths_list = traverse_dir_files_for_large(self.full_pdb_dir, ext="pdb")
all_pdb_path = self.all_pdb_format.format(len(paths_list))
print(f"[Info] 输出路径: {self.full_pdb_dir}")
write_list_to_file(all_pdb_path, paths_list)
print(f"[Info] 写入完成! {all_pdb_path}, 耗时: {time()-s_time}")
@staticmethod
def get_plddt_and_seq_once(pdb_path):
p = PDBParser(QUIET=True)
structure = p.get_structure('input', pdb_path)
plddt_list = []
for a in structure.get_residues():
b = a.get_unpacked_list()
if len(b) > 0:
plddt_list.append(b[0].get_bfactor())
plddt = np.average(np.array(plddt_list))
plddt = round(plddt, 4)
def_ids = ['I', 'Q', 'R', 'L', 'M', 'A', 'V', 'B', 'E', 'D', 'S', 'C',
'K', 'Y', 'X', 'J', 'T', 'F', 'G', 'W', 'H', 'P', 'Z', 'N']
d3to1 = d3to1_ex
seq_str_list = []
for model in structure:
for c_id, chain in enumerate(model):
seq = []
for residue in chain:
if residue.resname in d3to1.keys():
a = d3to1[residue.resname]
if a in def_ids:
seq.append(a)
else:
raise Exception(f"alpha not in default: {a}")
elif residue.resname == "HOH":
continue
if not seq:
continue
seq_str = "".join(seq)
seq_str_list.append(seq_str)
seq_str = seq_str_list[0]
return plddt, seq_str
@staticmethod
def parse_af_pdb(param):
"""
解析 AF PDB 文件
"""
pdb_path, idx = param
pdb_name = os.path.basename(pdb_path).split(".")[0]
plddt, seq = AfdbProcessor.get_plddt_and_seq_once(pdb_path)
seq_len = len(seq)
release_date = "2022-07-24" # v3
pdb_data = [pdb_name, plddt, release_date, seq, seq_len]
return pdb_data
def process_cluster_file(self, data_path):
"""
处理聚类文件
"""
print(f"[Info] 数据文件: {data_path}")
data_lines = read_file(data_path)
print(f"[Info] 数据行数: {len(data_lines)}")
file_name = os.path.basename(data_path).split(".")[0]
# 单进程
# pdb_data_list = []
# for pdb_name in data_lines:
# pdb_path = os.path.join(self.full_pdb_dir, pdb_name[:2], pdb_name)
# pdb_data = self.parse_af_pdb(pdb_path)
# pdb_data_list.append(pdb_data)
# column_names = ["pdb", "plddt", "release_date", "seq", "len"]
# df = pd.DataFrame(pdb_data_list, columns=column_names)
# out_csv = os.path.join(self.out_dir, f"{file_name}_labels_{len(pdb_data_list)}.csv")
# df.to_csv(out_csv)
# print(f"[Info] 写入完成: {out_csv}")
# 多进程
params_list = []
for idx, pdb_name in enumerate(data_lines):
pdb_path = os.path.join(self.full_pdb_dir, pdb_name[1:3], pdb_name)
params_list.append((pdb_path, idx))
# if idx == 50: # Debug
# break
pool = Pool(processes=40)
pdb_data_list = []
for res in list(tqdm(pool.imap(AfdbProcessor.parse_af_pdb, params_list), desc="[Info] pdb")):
pdb_data_list.append(res)
pool.close()
pool.join()
column_names = ["pdb", "plddt", "release_date", "seq", "len"]
df = pd.DataFrame(pdb_data_list, columns=column_names)
out_csv = os.path.join(self.out_dir, f"{file_name}_labels_{len(pdb_data_list)}.csv")
df.to_csv(out_csv)
print(f"[Info] 全部处理完成: {out_csv}")
def process(self):
self.process_cluster_file(self.pdb_name_path) # 处理文件
def main():
ap = AfdbProcessor()
ap.process()
if __name__ == '__main__':
main()
参考
- StackOverflow - Iterate over a very large number of files in a folder
- Convert a List to Pandas Dataframe (with examples)