万字长文介绍R package “vegan”——入门学习与重复文献数据
vegan介绍与入门
vegan是一个用于群落生态学(community ecology)分析的包,可以进行排序、多样性和差异性分析(ordination, diversity and dissimilarity)。
vegan包含了多样性分析、排序方法和差异性分析的工具。
示例一:unconstrained analysis(非约束排序)
### example1: unconstrained analysis
## NMDS
library(vegan)
data(varespec)
data("varechem")
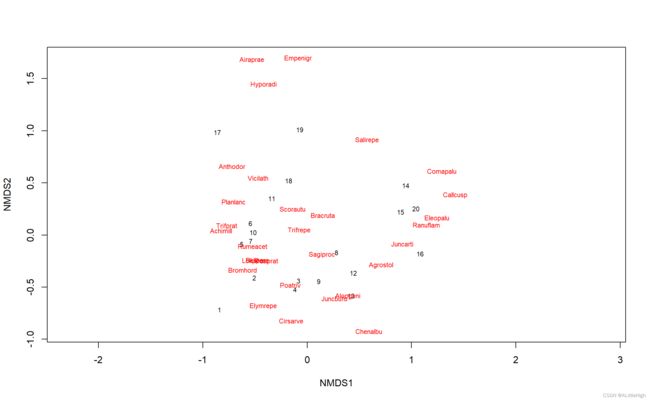
ord = metaMDS(varespec)
plot(ord, type = "t")
# fit environmental variables
ef = envfit(ord, varechem)
ef
plot(ef, p.max = 0.05)
> ef
***VECTORS
NMDS1 NMDS2 r2 Pr(>r)
N -0.05729 -0.99836 0.2536 0.035 *
P 0.61969 0.78484 0.1938 0.108
K 0.76642 0.64234 0.1809 0.109
Ca 0.68517 0.72838 0.4119 0.004 **
Mg 0.63250 0.77456 0.4270 0.002 **
S 0.19135 0.98152 0.1752 0.119
Al -0.87162 0.49019 0.5269 0.001 ***
Fe -0.93604 0.35190 0.4450 0.002 **
Mn 0.79872 -0.60171 0.5231 0.001 ***
Zn 0.61754 0.78654 0.1879 0.109
Mo -0.90308 0.42948 0.0609 0.526
Baresoil 0.92491 -0.38019 0.2508 0.045 *
Humdepth 0.93284 -0.36029 0.5200 0.001 ***
pH -0.64800 0.76164 0.2308 0.061 .
---
Signif. codes:
0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Permutation: free
Number of permutations: 999
示例二:constrainted analysis(约束排序)
### example2: constrained ordination (RDA)
## the example uses formula interface to define the modle
library(vegan)
data("dune")
data("dune.env")
# no constraints: PCA
mod0 = rda(dune ~ 1, dune.env)
mod0
plot(mod0)
# all environmental variables: Full model
mod1 = rda(dune ~ ., dune.env)
mod1
plot(mod1)
# automatic selection of variables by permutation P-values
mod = ordistep(mod0, scope=formula(mod1))
mod
plot(mod)
# permutation test for all variables
anova(mod)
# permutation test of "type Ⅲ" effects, or significance when a term is added to the model after all other terms
anova(mod, by = "margin")
# Plot only sample plots, use different symbols and draw SD ellipses for Managemenet classes
plot(mod, display = "sites", type = "n")
with(dune.env, points(mod, disp = "si", pch = as.numeric(Management)))
with(dune.env, legend("topleft", levels(Management), pch = 1:4, title = "Management"))
with(dune.env, ordiellipse(mod, Management, label = TRUE))
# add fitted surface of diversity to the model
ordisurf(mod, diversity(dune), add = TRUE)
示例三: analysis of dissimilarities(差异性分析)
### Example 3: analysis of dissimilarites a.k.a. non-parametric
### permutational anova
library(vegan)
data("dune")
data("dune.env")
adonis2(dune ~ ., dune.env)
adonis2(dune ~ Management + Moisture, dune.env)
文献数据复现
原文方法如下:… carried out NMDS with the metaMDS functioin and Gower’s Distance as the dissimilarity metric in the VEGAN package in R.
如此可知,我需要使用metaMDS函数并使用Gower’s Distance完成NMDS分析。
在操作前,我需要知道metaMDS函数和Gower’s Distance在vegan里分别是什么、如何使用?
metaMDS函数
Nonmetric Multidimensional Scaling with Stable Solution from Random Starts, Axis Scaling and Species Scores
大致意思是:通过随机开始、坐标轴缩放及物种评分来获得可靠的结果,这就是非度量多维测度。非度量多维测度是一种适用于非线性数据结构分析的复杂的迭代排序方法。在此不多描述。
metaMDS函数的用法
-
metaMDS(comm, distance = "bray", k = 2, try = 20, trymax = 20, engine = c("monoMDS", "isoMDS"), autotransform =TRUE, noshare = (engine == "isoMDS"), wascores = TRUE, expand = TRUE, trace = 1, plot = FALSE, previous.best, ...) -
plot(x, display = c("sites", "species"), choices = c(1, 2), type = "p", shrink = FALSE, ...) -
points(x, display = c("sites", "species"), choices = c(1,2), shrink = FALSE, select, ...) -
text(x, display = c("sites", "species"), labels, choices = c(1,2), shrink = FALSE, select, ...) -
scores(x, display = c("sites", "species"), shrink = FALSE, choices, tidy = FALSE, ...) -
metaMDSdist(comm, distance = "bray", autotransform = TRUE, noshare = TRUE, trace = 1, commname, zerodist = "ignore", distfun = vegdist, ...) -
metaMDSiter(dist, k = 2, try = 20, trymax = 20, trace = 1, plot = FALSE, previous.best, engine = "monoMDS", maxit = 200, parallel = getOption("mc.cores"), ...) -
initMDS(x, k=2) postMDS(X, dist, pc=TRUE, center=TRUE, halfchange, threshold=0.8, nthreshold=10, plot=FALSE, ...) -
metaMDSredist(object, ...)
我要使用的是第一个用法,下面了解以下metaMDS函数的参数
metaMDS函数的参数
| 参数 | 含义 |
|---|---|
| comm | 数据(community data) |
| distance | 差异性分析参数 |
| k | 维度数 |
| try,trymax | 随机迭代的最小和最大次数 |
| engine | 默认为monoMDS |
| autofransform | 如果没有群落数据,该项应设置为FALSE |
| noshare | TRUE/FLASE |
| wascross | 使用wascross函数计算物种分值 |
| expand | 在wascross函数中延伸物种的权重均值 |
| trace | 追踪函数 |
| plot | 图形化展示追踪过程:绘制中间过程 |
| previous.best | 从上一次结果处开始分析 |
| x | metaMDS结果 |
| choices | 轴线展示 |
| type | 图形类型:"p"表示点,"t"表示文本,"n"表示轴线 |
| display | 展示“sites”或“species” |
| shrink | 如果expand了,便会收缩 |
| tidy | 返回ggplot2可以使用的分值数据 |
| labels | 更改行名 |
| select | 挑选展示的条目 |
| X | 多维测度的配置 |
| commname | comm的名称 |
| zerodist | 零差异的处理:‘fail’、‘add’、‘ignore’ |
| distfun | 相异度函数 |
| maxit | 单次NMDS迭代的最大次数 |
| parallel | 并行线程 |
| dist | 相异度矩阵在多维标度中的应用 |
| pc | 旋转到主成分 |
| center | 中心配置 |
| halfchange | 将坐标轴刻度为半可变单位 |
| threshold | 在半变化缩放中使用的最大差异 |
| nthreshold | 半变化缩放中的最小点数 |
| object | 一个来自metaMDS的结果对象 |
metaMDS函数的官方示例
## The recommended way of running NMDS (Minchin 1987)
library(vegan)
data("dune")
## IGNORE_RDIFF_BEGIN
## Global NMDS using monoMDS
sol = metaMDS(dune)
sol
plot(sol, type = "t")
plot(sol, type = "p")
复现文献结果
数据准备
文件以上传,请自行下载。
NMDS分析
library(vegan)
library(openxlsx)
metrix = read.xlsx("D:/ALL_Softwares/R-4.2.0/library/vegan/2023Deanna.xlsx", 1,)
# tranform char to num, beacause xlse datatype are chars.
for (i in (2:ncol(metrix))){
metrix[,i] = as.numeric(metrix[,i])
}
# drop NAs
metrix = metrix[complete.cases(metrix),]
# drop nocl 1, which are speceis names
metrix= metrix[,2:ncol(metrix)]
# run NMDS analysis
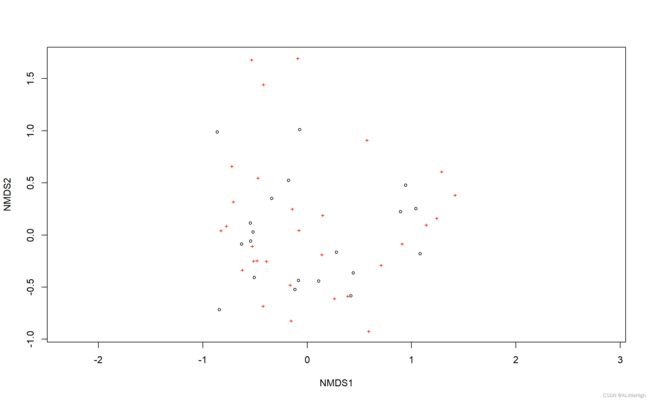
test = metaMDS(metrix, distance = 'gower', noshare = FALSE)
# draw result
plot(test)
stress 分析
# show stress analysis result
test$stress
> test$stress
[1] 0.1361019
P-values results
# calculate P-value
envfit(ord = test,env = metrix)
> envfit(ord = test,env = metrix)
***VECTORS
NMDS1
70001._Fruiting_calyx_length_(C1) -0.44058
70002._Fruiting_calyx_width_(C1) -0.42127
70003._Length_of_longest_fruiting_calyx_lobe_(C1) -0.04738
70021._Width_of_the_longest_fruiting_calyx_lobe_(C1) -0.15377
70010._Fruiting_calyx_base_invagination_(D1) -0.70729
70011._Fruiting_calyx_angled_(D1) -0.79430
70012._Fruiting_calyx_lobes_sinus_(D1) 0.16247
70014._Fruiting_calyx_primary_veins_distinct_from_other_vein_orders_(D1) -0.97775
70016._Fruiting_calyx_secondary_veins_distinct_from_other_vein_orders_and_emerge_from_base_(D1) -0.85812
70017._Fruiting_calyx_secondary_veins_fork_before_lobe_sinus_(D1) -0.91799
70018._Fruit_type_(D1) -0.78117
70019._Fruiting_calyx_inflated_(D1) -0.97198
70020._Fruiting_calyx_venation_pattern_(D1) -0.64975
NMDS2
70001._Fruiting_calyx_length_(C1) 0.89771
70002._Fruiting_calyx_width_(C1) 0.90694
70003._Length_of_longest_fruiting_calyx_lobe_(C1) 0.99888
70021._Width_of_the_longest_fruiting_calyx_lobe_(C1) 0.98811
70010._Fruiting_calyx_base_invagination_(D1) -0.70692
70011._Fruiting_calyx_angled_(D1) -0.60752
70012._Fruiting_calyx_lobes_sinus_(D1) -0.98671
70014._Fruiting_calyx_primary_veins_distinct_from_other_vein_orders_(D1) -0.20976
70016._Fruiting_calyx_secondary_veins_distinct_from_other_vein_orders_and_emerge_from_base_(D1) 0.51345
70017._Fruiting_calyx_secondary_veins_fork_before_lobe_sinus_(D1) 0.39661
70018._Fruit_type_(D1) 0.62431
70019._Fruiting_calyx_inflated_(D1) -0.23507
70020._Fruiting_calyx_venation_pattern_(D1) -0.76015
r2
70001._Fruiting_calyx_length_(C1) 0.2166
70002._Fruiting_calyx_width_(C1) 0.1170
70003._Length_of_longest_fruiting_calyx_lobe_(C1) 0.2824
70021._Width_of_the_longest_fruiting_calyx_lobe_(C1) 0.2508
70010._Fruiting_calyx_base_invagination_(D1) 0.4345
70011._Fruiting_calyx_angled_(D1) 0.4811
70012._Fruiting_calyx_lobes_sinus_(D1) 0.4865
70014._Fruiting_calyx_primary_veins_distinct_from_other_vein_orders_(D1) 0.1947
70016._Fruiting_calyx_secondary_veins_distinct_from_other_vein_orders_and_emerge_from_base_(D1) 0.7551
70017._Fruiting_calyx_secondary_veins_fork_before_lobe_sinus_(D1) 0.5930
70018._Fruit_type_(D1) 0.0752
70019._Fruiting_calyx_inflated_(D1) 0.5651
70020._Fruiting_calyx_venation_pattern_(D1) 0.0984
Pr(>r)
70001._Fruiting_calyx_length_(C1) 0.001
70002._Fruiting_calyx_width_(C1) 0.002
70003._Length_of_longest_fruiting_calyx_lobe_(C1) 0.001
70021._Width_of_the_longest_fruiting_calyx_lobe_(C1) 0.001
70010._Fruiting_calyx_base_invagination_(D1) 0.001
70011._Fruiting_calyx_angled_(D1) 0.001
70012._Fruiting_calyx_lobes_sinus_(D1) 0.001
70014._Fruiting_calyx_primary_veins_distinct_from_other_vein_orders_(D1) 0.001
70016._Fruiting_calyx_secondary_veins_distinct_from_other_vein_orders_and_emerge_from_base_(D1) 0.001
70017._Fruiting_calyx_secondary_veins_fork_before_lobe_sinus_(D1) 0.001
70018._Fruit_type_(D1) 0.001
70019._Fruiting_calyx_inflated_(D1) 0.001
70020._Fruiting_calyx_venation_pattern_(D1) 0.001
70001._Fruiting_calyx_length_(C1) ***
70002._Fruiting_calyx_width_(C1) **
70003._Length_of_longest_fruiting_calyx_lobe_(C1) ***
70021._Width_of_the_longest_fruiting_calyx_lobe_(C1) ***
70010._Fruiting_calyx_base_invagination_(D1) ***
70011._Fruiting_calyx_angled_(D1) ***
70012._Fruiting_calyx_lobes_sinus_(D1) ***
70014._Fruiting_calyx_primary_veins_distinct_from_other_vein_orders_(D1) ***
70016._Fruiting_calyx_secondary_veins_distinct_from_other_vein_orders_and_emerge_from_base_(D1) ***
70017._Fruiting_calyx_secondary_veins_fork_before_lobe_sinus_(D1) ***
70018._Fruit_type_(D1) ***
70019._Fruiting_calyx_inflated_(D1) ***
70020._Fruiting_calyx_venation_pattern_(D1) ***
---
Signif. codes:
0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Permutation: free
Number of permutations: 999
后记
因为NMDS采用了随机计算,所以每次结果都不同,也无法重现文献结果,但大致结果相同。
本文中,我已经学会并详细记录了使用vegan进行分类(对物种进行排序分类),工作流程包括使用metaMDS函数、stress分析和P-values结果。
纰漏:唯一无法实现(或者说我对vegan理解的不到位),原始文献中:All characters were treated as symmetric except fruit type, which was considered ordered. 也就是说,原始文献中进行NMDS分析时,对数据进行了处理:果实类型处理为orderd(排序的),除此以外均处理为symmetric(对称的)。
另外,非常感谢王翰臣给予的帮助,非常感谢!