VAE, the principle and the code
VAE
keywords encoder, decoder
文章目录
- VAE
-
- Concepts
-
- Facts
- Assumptions
- variational inference(VI)
- variational autoencoder(AVE) architecture
-
- architecture
- Bayes-by-BP
- PCA/SVD as a VAE
- Basic Model
- Full Model
Concepts
Variational autoencoder(VAE) is a latent model p ( x , z ∣ θ ) p(x, z|\theta) p(x,z∣θ).
probabilistic encoder-decoder:
-
probabilistic decoder/likelihood/generating proba: p ( x ∣ z , θ ) p(x|z,\theta) p(x∣z,θ), the distribution of the decoded variable given the encoded one,
-
probabilistic encoder/posterior/predicting(discriminant, respondant) proba/encoding: p ( z ∣ x , θ ) p(z|x,\theta) p(z∣x,θ), the distribution of the encoded variable given the decoded one.
graph: z → x ( ← θ ) z\to x (\leftarrow \theta) z→x(←θ)
Task: to find q ( z ) ≈ p ( z ∣ x ) q(z)\approx p(z|x) q(z)≈p(z∣x), or denoted as q ( z ∣ x ) q(z|x) q(z∣x) to emphsize the dependence on x x x. (variational inference)
loss: D K L ( q ( z ) ∥ p ( z ∣ x ) ) D_{KL}(q(z)\|p(z|x)) DKL(q(z)∥p(z∣x))
likelihood: l ( θ ) = ∑ x ln ∫ z p ( x ∣ z , θ ) p ( z ∣ θ ) d z l(\theta)=\sum_x\ln\int_z p(x|z,\theta) p(z|\theta)dz l(θ)=∑xln∫zp(x∣z,θ)p(z∣θ)dz
Variational Lower Bound/Evidence Lower Bound(ELBO)
L x : = E z ∼ q ( log p ( x , z ) − log q ( z ) ) = Q ( p , q ) + H ( q ) ( = F ( p , q ) ) L_x := E_{z\sim q}(\log p(x,z)-\log q(z))\\ = Q(p,q)+H(q) (=F(p,q)) Lx:=Ez∼q(logp(x,z)−logq(z))=Q(p,q)+H(q)(=F(p,q))
where q q q alwayes depends on x x x.
Remark L x L_x Lx is also called free energy.
ELBO = expected likelihood + entropy
(EM algo. / MM algo.)
Facts
Identity
variational inequality
log p ( x ) = D K L ( q ( z ) ∥ p ( z ∣ x ) ) + L x ≥ L x \log p(x) = D_{KL}(q(z)\| p(z|x))+L_x\geq L_x logp(x)=DKL(q(z)∥p(z∣x))+Lx≥Lx
remark likelihood = divergence + ELBO(free energy)
L x = E z ∼ q ( log p ( x ∣ z ) ) − D K L ( q ( z ) ∥ p Z ( z ) ) , L_x=E_{z\sim q}(\log p(x|z))-D_{KL}(q(z)\|p_Z(z)), Lx=Ez∼q(logp(x∣z))−DKL(q(z)∥pZ(z)),
ELBO = Reconstruction loss + Regularization term
Reconstruction loss: − E z ∼ q ( log p ( x ∣ z ) ) -E_{z\sim q}(\log p(x|z)) −Ez∼q(logp(x∣z))
Regularization term: D K L ( q ∥ p Z ) D_{KL}(q\| p_Z) DKL(q∥pZ)
For samples D D D,
L D = ∑ x ∈ D E z ∼ q log p ( x ∣ z ) − D K L ( q ∥ p Z ) L_D = \sum_{x\in D}E_{z\sim q}\log p(x|z) - D_{KL}(q\|p_Z) LD=x∈D∑Ez∼qlogp(x∣z)−DKL(q∥pZ)
remark For fixed p p p, min p , q D K L ⟺ max p , q L x \min_{p,q} D_{KL} \iff \max_{p,q} L_x minp,qDKL⟺maxp,qLx.
diff. between EM and VAE
- EM algorithm: solve max L x \max L_x maxLx by coordinate ascent
- VAE: by SGD
Parameter form
L D ( θ , ϕ ) = ∑ x ∈ D ( E z ∼ q x , ϕ log p ( x ∣ z , θ ) − D K L ( q ( z ∣ x , ϕ ) ∥ p ( z ∣ θ ) ) ) = ∑ x ∈ D ( E z ∼ q x , ϕ log p ( x ∣ z , θ ) − D K L ( q ( z ∣ x , ϕ ) ∥ p ( z ) ) ) ( i f θ → x ← z ) L_D(\theta,\phi) = \sum_{x\in D}(E_{z\sim q_{x,\phi}}\log p(x|z,\theta) - D_{KL}(q(z|x,\phi)\|p(z|\theta)))\\ =\sum_{x\in D}(E_{z\sim q_{x,\phi}}\log p(x|z,\theta) - D_{KL}(q(z|x,\phi)\|p(z))) ~~(if~\theta \to x \leftarrow z) LD(θ,ϕ)=x∈D∑(Ez∼qx,ϕlogp(x∣z,θ)−DKL(q(z∣x,ϕ)∥p(z∣θ)))=x∈D∑(Ez∼qx,ϕlogp(x∣z,θ)−DKL(q(z∣x,ϕ)∥p(z))) (if θ→x←z)
where variational parameters: ϕ \phi ϕ, generative parameters: θ \theta θ.
Assumptions
Gaussian assumption: p ( z ) , p ( x ∣ z ) p(z),p(x|z) p(z),p(x∣z): Gaussian distr.
Z ∼ N ( 0 , 1 ) , X ∣ Z = z ∼ N ( f ( z ) , c ) , f ∈ F , c > 0. Z\sim N(0,1),\\ X|Z=z\sim N(f(z),c),f\in F, c>0. Z∼N(0,1),X∣Z=z∼N(f(z),c),f∈F,c>0.
It is intractable to compute p ( z ∣ x ) p(z|x) p(z∣x) by Bayesian formula.
variational inference(VI)
VI is a technique to approximate complex distributions
Continue from (4)
We are going to approximate p ( z ∣ x ) p(z|x) p(z∣x) by a Gaussian distribution q x ( z ) q_x(z) qx(z) whose mean and covariance are defined by two functions, g g g and h h h, of the parameter x x x.
q ( Z ∣ x ) ∼ N ( g ( x ) , h ( x ) ) , g ∈ G , h ∈ H , q(Z|x)\sim N(g(x),h(x)), g\in G, h\in H, q(Z∣x)∼N(g(x),h(x)),g∈G,h∈H,
where variational parameter ϕ = ( g , h ) ∈ G × H \phi=(g, h)\in G\times H ϕ=(g,h)∈G×H.
Fixed f f f (hence log p ( x ) \log p(x) logp(x) is a constant), solve the following optimialization problem,
max L x ⟺ min g , h D K L ( q x ( z ) ∥ p ( z ∣ x ) ) ≃ min g , h − E log p ( x ∣ z ) + D K L ( q x ∥ p Z ) = min g , h 1 2 c E ∥ x − f ( z ) ∥ 2 + D K L ( q x ∥ p Z ) . \max L_x \iff \min_{g,h} D_{KL}(q_x(z)\|p(z|x))\\ \simeq \min_{g,h} -E\log p(x|z) + D_{KL}(q_x\|p_Z)\\ =\min_{g,h} \frac{1}{2c}E\|x-f(z)\|^2 + D_{KL}(q_x\| p_Z). maxLx⟺g,hminDKL(qx(z)∥p(z∣x))≃g,hmin−Elogp(x∣z)+DKL(qx∥pZ)=g,hmin2c1E∥x−f(z)∥2+DKL(qx∥pZ).
Then find optimal f f f,
max f E z ∼ N ( g ( x ) , h ( x ) ) log p ( x ∣ z ) = min f E z ∥ x − f ( z ) ∥ 2 \max_f E_{z\sim N(g(x),h(x))}\log p(x|z)=\min_f E_z\|x-f(z)\|^2 fmaxEz∼N(g(x),h(x))logp(x∣z)=fminEz∥x−f(z)∥2
===>
= 1 2 c min f , g , h E z ∥ x − f ( z ) ∥ 2 + D K L ( q x ∥ p Z ) . = \frac{1}{2c}\min_{f,g,h}E_z\|x-f(z)\|^2 + D_{KL}(q_x\| p_Z). =2c1f,g,hminEz∥x−f(z)∥2+DKL(qx∥pZ).
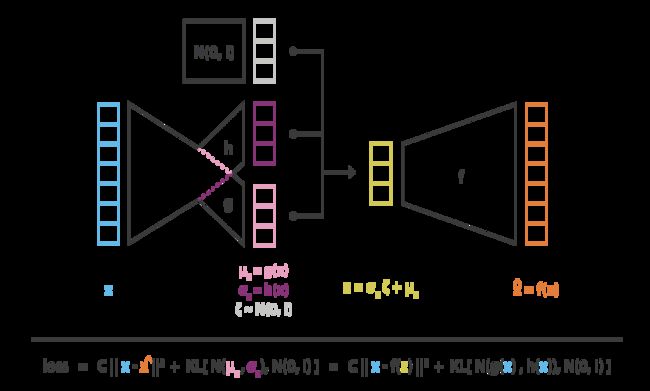
variational autoencoder(AVE) architecture
Based on Gaussian assumption.
architecture
encoder: x → z = g ( x ) + h ( x ) ζ ∼ N ( g ( x ) , h ( x ) ) x\to z=g(x)+h(x)\zeta\sim N(g(x),h(x)) x→z=g(x)+h(x)ζ∼N(g(x),h(x)) and ζ ∼ N ( 0 , 1 ) \zeta\sim N(0,1) ζ∼N(0,1) (reparameterization), as an approximation of p ( z ∣ x ) p(z|x) p(z∣x)
decoder: z → x ∼ N ( f ( z ) , c ) z\to x\sim N(f(z),c) z→x∼N(f(z),c)
reparametrisation trick
L ( x ) = C E z ∥ x − f ( z ) ∥ 2 + D K L ( N ( g ( x ) , h ( x ) ) ∥ N ( 0 , 1 ) ) L(x)=CE_z\|x-f(z)\|^2 + D_{KL}(N(g(x),h(x))\| N(0,1)) L(x)=CEz∥x−f(z)∥2+DKL(N(g(x),h(x))∥N(0,1))
where D K L D_{KL} DKL of the diagonal normal distr. and standard normal distr. is D K L ( D N , S N ) : = 1 2 ∑ i = 1 k ( σ i 2 + μ i 2 − 1 − ln σ i 2 ) D_{KL}(DN, SN):=\frac{1}{2}∑_{i=1}^k(σ^2_i+μ^2_i−1−\ln σ^2_i) DKL(DN,SN):=21∑i=1k(σi2+μi2−1−lnσi2), where μ i = g i ( x ) , σ i 2 = h i ( x ) \mu_i=g_i(x),\sigma_i^2=h_i(x) μi=gi(x),σi2=hi(x).
see Wiki of KL divergence.
Bayes-by-BP
Algo
- initalize ϕ ← ϕ 0 \phi\leftarrow \phi_0 ϕ←ϕ0
- loop from i=0 to N
- z ∼ q ( z ∣ x , ϕ ) z\sim q(z|x,\phi) z∼q(z∣x,ϕ);
- θ \theta θ maximizes log p ( x ∣ z , θ ) \log p(x|z,\theta) logp(x∣z,θ);
- calculate ELBO L ( θ , ϕ ) L(\theta,\phi) L(θ,ϕ);
- update ϕ \phi ϕ by GD;
General form of Algorithm
opt F ( θ ) = E z ∼ q θ f ( θ , z ) F(\theta)=E_{z\sim q_{\theta}}f(\theta,z) F(θ)=Ez∼qθf(θ,z):
- guess θ \theta θ, generate z ∼ q θ z\sim q_\theta z∼qθ;
- Do GD for f ( θ , z ) f(\theta,z) f(θ,z) to update θ \theta θ;
PCA/SVD as a VAE
General form encoder-decoder model
min d , e ∣ x − d ( e ( x ) ) ∣ \min_{d,e}|x-d(e(x))| d,emin∣x−d(e(x))∣
SVD, dim of data space = p, laten space = q
min V ∣ X − X V q V q ′ ∣ V : O ( p ) \min_{V}|X-X V_q V_q'|\\ V:O(p) Vmin∣X−XVqVq′∣V:O(p)
xi ~ N(0,1)
\
D -- > Z --> X = ZV'
where Z ∼ N ( 0 , D ) , X = Z V ′ Z\sim N(0,D),X=ZV' Z∼N(0,D),X=ZV′.
stat. model of encoder-decoder
- encoder: p ( x ∣ z ) p(x|z) p(x∣z)
- decoder: p ( z ∣ x ) p(z|x) p(z∣x)
as the conditional proba. of p ( x , z ) p(x,z) p(x,z)
generating rask: z ∼ N ( 0 , 1 ) ⇒ x ∼ p ( x ∣ z ) z\sim N(0,1) \Rightarrow x\sim p(x|z) z∼N(0,1)⇒x∼p(x∣z)
Basic Model
graph: c -> z -> x (<-theta), where c c c: context.
Likelihood: p ( D ∣ θ ) = ∏ x ∈ D ∫ p ( x ∣ z , θ ) p ( z ∣ c , θ ) d z p(D|\theta)=\prod_{x\in D}\int p(x|z,\theta)p(z|c,\theta) dz p(D∣θ)=∏x∈D∫p(x∣z,θ)p(z∣c,θ)dz
variational lower bound:
L D = E c ∼ q D ( L D ∣ c ) − D K L ( q D ∥ p c ) L_D = E_{c\sim q_D}(L_{D|c})-D_{KL}(q_D\|p_c) LD=Ec∼qD(LD∣c)−DKL(qD∥pc)
Full Model
graph: c -> z1,...,zT -> x (<-theta), where c c c: context.
References
understanding-variational-autoencoders
Ming Ding. The road from MLE to EM to VAE: A brief tutorial,2022.
H Edwards, A. Storkey. Towards a neural statistician,2017.
Codes
code on line:
VAE-keras
my code:
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
VAE
"""
import numpy as np
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
class Sampling(layers.Layer):
"""Sampling Layer for reparameterization
Uses (z_mean, z_log_var) to sample z, the vector encoding a digit.
"""
def call(self, inputs):
z_mean, z_log_var = inputs
batch, dim = tf.shape(z_mean)[0], tf.shape(z_mean)[1]
epsilon = tf.keras.backend.random_normal(shape=(batch, dim))
return z_mean + tf.exp(0.5 * z_log_var) * epsilon
class VAE(keras.Model):
# architechture of VAE
def __init__(self, latent_dim, **kwargs):
super().__init__(**kwargs)
self.latent_dim = latent_dim
self.total_loss_tracker = keras.metrics.Mean(name="total_loss")
self.reconstruction_loss_tracker = keras.metrics.Mean(name="reconstruction_loss")
self.kl_loss_tracker = keras.metrics.Mean(name="kl_loss")
@property
def metrics(self):
return [
self.total_loss_tracker,
self.reconstruction_loss_tracker,
self.kl_loss_tracker,
]
def train_step(self, data):
with tf.GradientTape() as tape:
z_mean, z_log_var, z = self.encoder(data)
reconstruction = self.decoder(z)
reconstruction_loss = tf.reduce_mean(
tf.reduce_sum(
keras.losses.binary_crossentropy(data, reconstruction), axis=(1, 2)
)
) # Hb(x_i, f(z_i)) z_i ~ N(g(x_i),h(x_i))
kl_loss = 0.5 * tf.reduce_sum(tf.square(z_mean) + tf.exp(z_log_var) - 1 - z_log_var, axis=1)
kl_loss = tf.reduce_mean(kl_loss) # KL-div of N and SN
total_loss = reconstruction_loss + kl_loss
grads = tape.gradient(total_loss, self.trainable_weights)
self.optimizer.apply_gradients(zip(grads, self.trainable_weights))
self.total_loss_tracker.update_state(total_loss)
self.reconstruction_loss_tracker.update_state(reconstruction_loss)
self.kl_loss_tracker.update_state(kl_loss)
return {
"loss": self.total_loss_tracker.result(),
"reconstruction_loss": self.reconstruction_loss_tracker.result(),
"kl_loss": self.kl_loss_tracker.result()}
@classmethod
def make_from_data(cls, X, latent_dim, *args, **kwargs):
shape = X.shape[1:3]
return cls.make(shape, latent_dim, *args, **kwargs)
@classmethod
def make(cls, shape, latent_dim, *args, **kwargs):
assert len(shape)>=2, "Sorry, this VAE works only for images; the ndim of each sample>=2!"
model = cls(latent_dim, *args, **kwargs)
model.encoder = cls.make_encoder(shape, latent_dim)
model.decoder = cls.make_decoder(shape, latent_dim)
model.compile(optimizer=keras.optimizers.Adam())
return model
@classmethod
def make_encoder(cls, shape, latent_dim):
# shape: the shape of input data
if len(shape)==2:
height, width = shape
n_channels = 1
elif len(shape)==3:
height, width, n_channels = shape
encoder_inputs = layers.Input(shape=(height, width, n_channels))
x = layers.Conv2D(32, 3, activation="relu", strides=2, padding="same")(encoder_inputs)
x = layers.Conv2D(64, 3, activation="relu", strides=2, padding="same")(x)
x = layers.Flatten()(x)
x = layers.Dense(16, activation="relu")(x)
z_mean = layers.Dense(latent_dim, name="z_mean")(x) # g(x)
z_log_var = layers.Dense(latent_dim, name="z_log_var")(x) # h(x)
z = Sampling()([z_mean, z_log_var]) # z ~ N(g(x),h(x))
return keras.Model(encoder_inputs, [z_mean, z_log_var, z], name="encoder")
@classmethod
def make_decoder(cls, shape, latent_dim):
# x = f(z), shape: the shape of the output of the decoder
if len(shape)==2:
height, width = shape
n_channels = 1
elif len(shape)==3:
height, width, n_channels = shape
small_height, small_width = height // 4, width // 4
decoder = keras.Sequential(name="decoder") # f(z)
latent_inputs = keras.Input(shape=(latent_dim,))
decoder.add(latent_inputs)
decoder.add(layers.Dense(small_height * small_width * 64, activation="relu"))
decoder.add(layers.Reshape((small_height, small_width, 64)))
decoder.add(layers.Conv2DTranspose(64, 3, activation="relu", strides=2, padding="same"))
decoder.add(layers.Conv2DTranspose(32, 3, activation="relu", strides=2, padding="same"))
decoder.add(layers.Conv2DTranspose(1, 3, activation="sigmoid", padding="same"))
return decoder
def trainsform(self, X):
M, _, _ = self.encoder(X)
return M
def inverse_trainsform(self, Z):
return self.decoder.predict(Z)
def plot_latent_space(vae, n=11, *args, **kwargs):
import itertools
import matplotlib.pyplot as plt
# display a n*n 2D manifold of digits
scale = 1.0
# linearly spaced coordinates corresponding to the 2D plot
# of digit classes in the latent space
grid_x = np.linspace(-scale, scale, n)
grid_y = np.linspace(-scale, scale, n)[::-1]
z_sample = np.hstack((list(itertools.product(grid_y, grid_x)), np.random.normal(size=(n**2, vae.latent_dim-2))))
x_decoded = vae.inverse_trainsform(z_sample)
figure = np.block([[x_decoded[i*n+j].reshape((width, height)) for j in range(n)] for i in range(n)])
plt.figure(*args, **kwargs)
start_range_x, start_range_y = width // 2, height // 2
end_range_x = n * width + start_range_x
end_range_y = n * height + start_range_y
pixel_range_x = np.arange(start_range_x, end_range_x, width)
pixel_range_y = np.arange(start_range_y, end_range_y, height)
sample_range_x = np.round(grid_x, 1)
sample_range_y = np.round(grid_y, 1)
plt.xticks(pixel_range_x, sample_range_x)
plt.yticks(pixel_range_y, sample_range_y)
plt.xlabel("$z_0$")
plt.ylabel("$z_1$")
plt.imshow(figure, cmap="Greys_r")
plt.show()
if __name__ == '__main__':
# from get_hanzi import X_train
# input your data here, images in size of 4m X 4n
X_train /= 255
X_train = X_train>0.5 # binarize
X_train = np.expand_dims(X_train, -1).astype("float32")
vae = VAE.make_from_data(X_train, latent_dim=15)
vae.fit(X_train, epochs=200, batch_size=16, verbose=False)
plot_latent_space(vae)