Deep Learning With Pytorch - 数据预处理,以导入LUNA16数据集为例
文章目录
-
- 数据集简介
-
- 什么是CT扫描?
- 导入大型数据集并不是一份轻松的工作
- 在Jupyter Notebook中导入LUNA16数据集
-
- 导入可能用到的第三方库:
- LUNA16存放路径:
- 用 pandas 读取 candidates.csv;
- 读取 annotations.csv
- 导入subset0和subset1的.mhd
- 剔除路径、后缀名
- 创建字典diameter_dict:
- 创建candidateInfo_list:
- 打印一张CT扫描数据
- 数据类型转换,将IRC转为XYZ
- 从CT扫描中取出⼀个结节
- 定义getCandidateInfoList
- 封装成类的完整程序:
数据集简介
该数据集来源于LUNA16挑战赛,该数据集包括来自 LIDC/IDRI 数据集的不同格式的CT扫描图像以及附加注释。
The publicly available LIDC/IDRI database. This data uses the Creative Commons Attribution 3.0 Unported License. We excluded scans with a slice thickness greater than 2.5 mm. In total, 888 CT scans are included. The LIDC/IDRI database also contains annotations which were collected during a two-phase annotation process using 4 experienced radiologists. Each radiologist marked lesions they identified as non-nodule, nodule < 3 mm, and nodules >= 3 mm. See this publication for the details of the annotation process. The reference standard of our challenge consists of all nodules >= 3 mm accepted by at least 3 out of 4 radiologists. Annotations that are not included in the reference standard (non-nodules, nodules < 3 mm, and nodules annotated by only 1 or 2 radiologists) are referred to as irrelevant findings. The list of irrelevant findings is provided inside the evaluation script (annotations_excluded.csv).
下载地址
我们的CT数据来⾃2个文件:⼀个.mhd⽂件包含元数据头信息,另⼀个.raw文件包含组成三维数组的原始数据。我们所讨论的CT扫描的每个⽂件的名称都以⼀个称为系列UID的唯⼀标识符开始,该名称
依据医学中的数字成像和通信(DICOM)命名法。例如,对于uid1.2.3系列,有2个文件——1.2.3.mhd和1.2.3.raw.
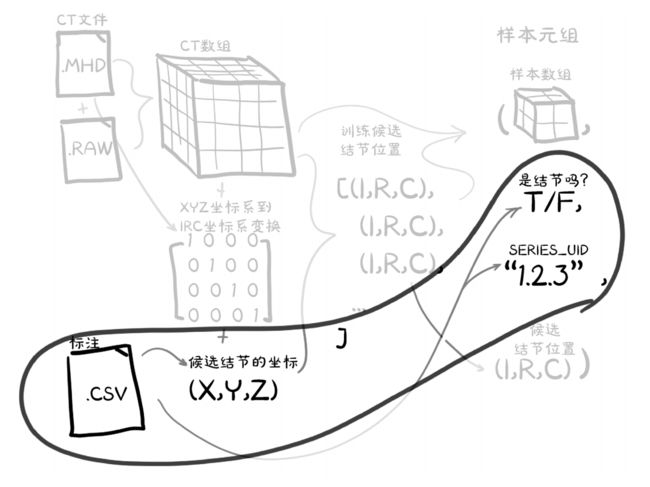
图中的 (I,R,C) 代表(索引,行,列),之后我们会介绍相应的功能函数实现 (X,Y,Z) 和 (I,R,C) 之间的相互转换。
什么是CT扫描?
CT扫描本质上是三维X射线,以单通道数据(灰度)的三维数组表示,也就是一组堆叠的灰度PNG图像:
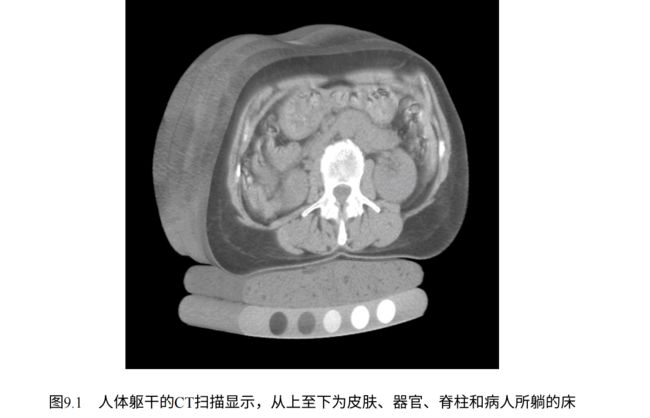
CT扫描(Computed Tomography),即计算机断层扫描,实际上利用特殊仪器测量的放射性密度(成像原理参考Radon变换/逆变换),它具备检查材料的质量密度和原子序数的功能。就我们的目的而言,这种区别无关紧要,因为无论输入的确切单位是什么,模型都将使用并从CT数据中学习。
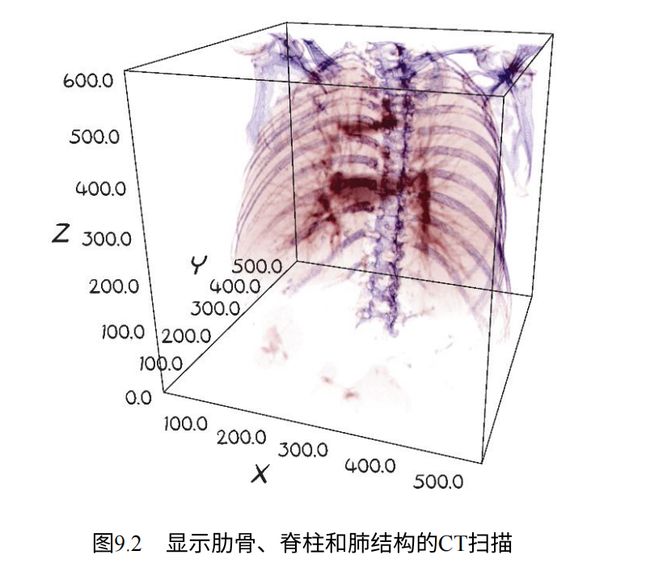
这种三维显示还允许我们通过隐藏我们不感兴趣的组织类型来查看物体内部。例如,我们可以以三维方式呈现数据,并将可见性限制为仅骨骼和肺组织:
导入大型数据集并不是一份轻松的工作
对于很多初学者来说,接触到的数据集如果是一组.csv或.xls,那么在Python中通过pandas就可以很方便地读入工程中查看,之后就是选取特征数和batch_size来作为模型输入了。而对于一些大型的数据集而言,尤其是随着数据的维度上升,从一维的时间序列到二维、多channels的图像数据,再到高维度的视频信号,这时的数据预处理并不是轻松几行代码就能完成的工作。
以下是根据一段Pytorch官方给出的数据导入范例,记录的学习笔记,在没有读过Deep Learning With Pytorch这本书的前提下,原本简单易懂的文章,本文会使读者看了觉得更懵逼,所以还是建议先阅读原书9-11章,github上有完整工程;
官方代码非常精简实用,对python中常见的数据结构,以及np.array,tensor等数据的读取、转换操作十分流畅,很适合作为学习资料。
在Jupyter Notebook中导入LUNA16数据集
导入可能用到的第三方库:
import copy
import csv
import functools
import glob
import os
import SimpleITK as sitk
import numpy as np
import pandas as pd
import collections
from collections import namedtuple
LUNA16存放路径:
可以自定义路径方便读取,我的是放在:
path = 'D:/papers/dataset/LUNA16/'
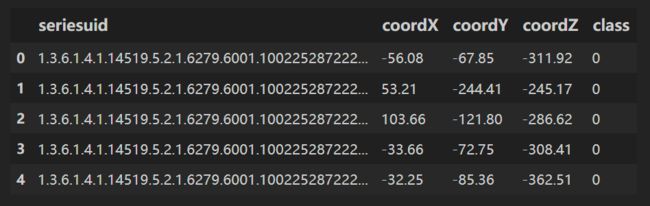
用 pandas 读取 candidates.csv;
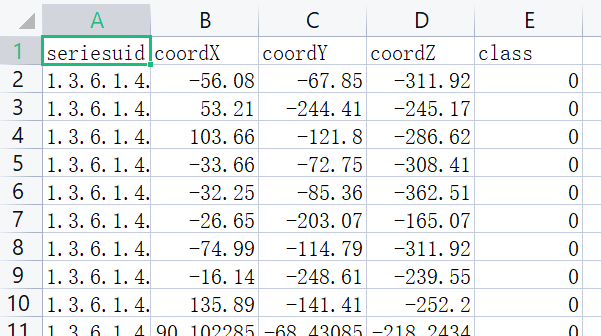
candidates.csv⽂件中的LUNA标注包含了CT序列、候选结节的位置,以及⼀个标识(指示该候选者是否真的是结节)
df = pd.read_csv(path+'candidates.csv')
df.head()
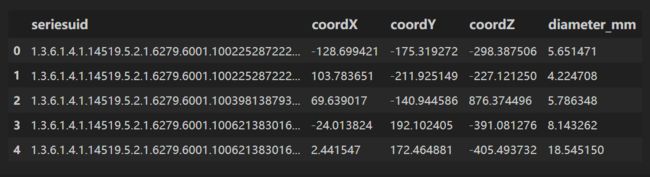
读取 annotations.csv
annotations.csv⽂件包含⼀些已标注为实际结节的候选者的信息,我们对diameter_mm(结节直径)列所代表的信息特别感兴趣。
annotations = pd.read_csv(path+'annotations.csv')
annotations.head()
导入subset0和subset1的.mhd
我们的CT数据来⾃2个⽂件:⼀个.mhd⽂件包含元数据头信息,另⼀个.raw⽂件包含组成三维数组的原始数据。
LUNA16中有 subset0-subset4 约35G的CT扫描影像数据(完整的数据集包含subset0~9约200多G的数据),本文仅选取了subset0和subset1;
我们所讨论的CT扫描的每个⽂件的名称都以⼀个称为系列UID的唯⼀标识符开始,该名称依据医学中的数字成像和通信(DICOM)命名法。例如,对于uid1.2.3系列,有2个⽂件——1.2.3.mhd和1.2.3.raw
mhd_list = glob.glob(path+'subset*/*.mhd')
功能解析:
glob.glob 接受一个字符串参数,该参数可以包含通配符字符(例如 * 和 ?)以及路径分隔符,然后它会在指定的路径中搜索与模式匹配的文件路径,并将匹配的文件路径返回为一个列表。
以下是一个示例,演示了如何使用 glob.glob 来匹配某个目录下的所有 .txt 文件:
import glob
txt_files = glob.glob('/path/to/directory/*.txt')
print(txt_files)
剔除路径、后缀名
从名为mhd_list的列表中,针对每个文件路径,提取文件名的部分(去掉扩展名),然后将这些部分组成一个集合presentOnDisk_set:
presentOnDisk_set = {os.path.split(p)[-1][:-4] for p in mhd_list}
功能解析:
os.path.split(mhd_list[0]),os.path.split(mhd_list[0])[1],os.path.split(mhd_list[0])[1][:-4]
打印结果:
(('D:/papers/dataset/LUNA16\\subset0',
'1.3.6.1.4.1.14519.5.2.1.6279.6001.105756658031515062000744821260.mhd'),
'1.3.6.1.4.1.14519.5.2.1.6279.6001.105756658031515062000744821260.mhd',
'1.3.6.1.4.1.14519.5.2.1.6279.6001.105756658031515062000744821260')
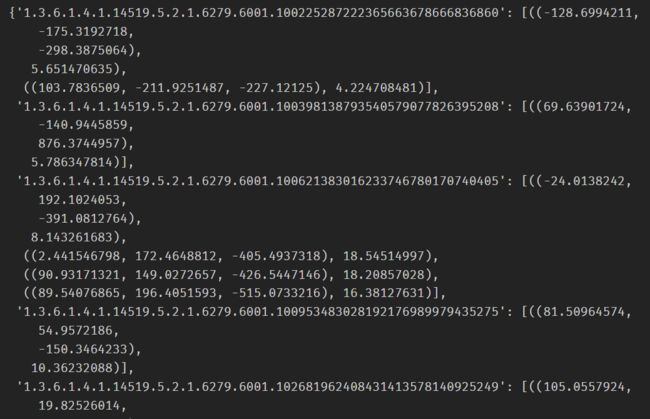
创建字典diameter_dict:
diameter_dict = {}
with open(path+'annotations.csv', "r") as f:
for row in list(csv.reader(f))[1:]:
series_uid = row[0]
annotationCenter_xyz = tuple([float(x) for x in row[1:4]])
annotationDiameter_mm = float(row[4])
diameter_dict.setdefault(series_uid, []).append(
(annotationCenter_xyz, annotationDiameter_mm)
)
功能解析:
把完整的每个 row 都打印出来看一下,发现就是一组组 List,所以 row[0] 就代表了第一列的 series_uid,row[1:4] 分别是位置坐标和 diameter :
创建candidateInfo_list:
与diameter_dict类似,candidateInfo_list从candidates.csv中获取series_uid,center_xyz坐标信息,并存放在一个Tuple里,最终分类嵌入candidateInfo_list;
requireOnDisk_bool = True
CandidateInfoTuple = namedtuple(
'CandidateInfoTuple',
'isNodule_bool, diameter_mm, series_uid, center_xyz',
)
candidateInfo_list = []
with open(path+'candidates.csv', "r") as f:
for row in list(csv.reader(f))[1:]: # 从第二行开始,第一行[0]是标题跳过
series_uid = row[0]
if series_uid not in presentOnDisk_set and requireOnDisk_bool:
continue
isNodule_bool = bool(int(row[4]))
candidateCenter_xyz = tuple([float(x) for x in row[1:4]])
candidateDiameter_mm = 0.0
for annotation_tup in diameter_dict.get(series_uid, []):
annotationCenter_xyz, annotationDiameter_mm = annotation_tup
for i in range(3):
delta_mm = abs(candidateCenter_xyz[i] - annotationCenter_xyz[i])
if delta_mm > annotationDiameter_mm / 4:
break
else:
candidateDiameter_mm = annotationDiameter_mm
break
candidateInfo_list.append(CandidateInfoTuple(
isNodule_bool,
candidateDiameter_mm,
series_uid,
candidateCenter_xyz,
))
功能解析:通常在python中用idx表示行,用row表示列,而在matlab中row代表行,column代表列。
import csv
with open('your_csv_file.csv', 'r') as f:
csv_reader = csv.reader(f)
for idx, row in enumerate(csv_reader):
if 2 <= idx <= 6: # 读取第3到第7行的数据
data = row[1] # 第二列的数据
# 在这里处理 data,它是第二列的数据
if idx > 6: # 已读取完第7行,结束循环
break
这段代码表示读入csv数据的第3-7行,第2列的所有数据。
打印一张CT扫描数据
series_uid = '1.3.6.1.4.1.14519.5.2.1.6279.6001.100684836163890911914061745866'
mhd_path = glob.glob(
path+'subset*/{}.mhd'.format(series_uid)
)[0]
ct_mhd = sitk.ReadImage(mhd_path)
ct_a = np.array(sitk.GetArrayFromImage(ct_mhd), dtype=np.float32)
ct_a.shape 为:(171, 512, 512),代表(I,R,C),即每张CT的截面图都是512x512的,series_uid = '1.3.6.1.4.1.14519.5.2.1.6279.6001.100684836163890911914061745866’中包含171张截面图。
我们可以尝试打印其中几张CT扫描截面图:
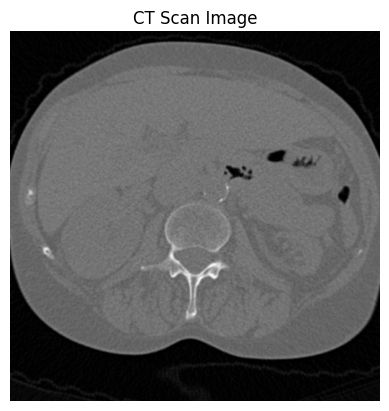
import matplotlib.pyplot as plt
# 显示CT图像
plt.imshow(ct_a[0], cmap='gray') # 显示第一个切片
plt.axis('off') # 关闭坐标轴
plt.title('CT Scan Image')
plt.show()
ct_a[10]:
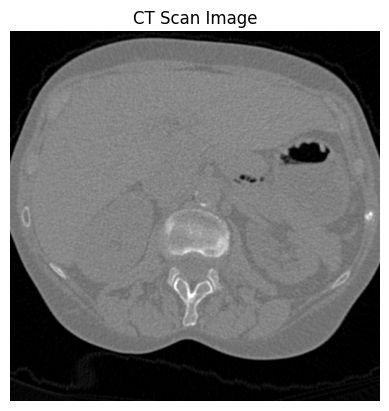
ct_a[50]:
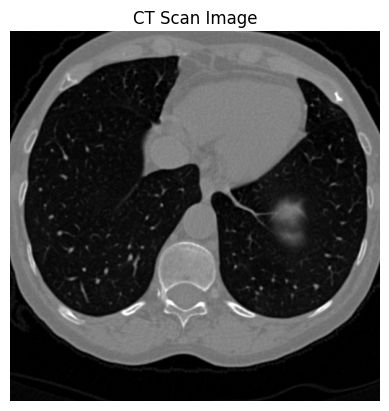
ct_a[170]:
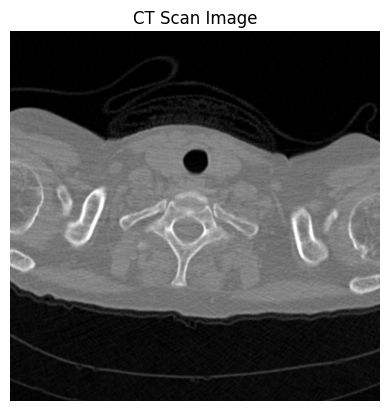
数据类型转换,将IRC转为XYZ
import util # 调用pytorch范例中自带的另一个.py
util.irc2xyz
ct_a.clip(-1000, 1000, ct_a) # 将像素值限制在 -1000 到 1000 的范围内
series_uid = series_uid
hu_a = ct_a # 在做CT检查时,Hu是反映人体各组织的密度的指标
IrcTuple = collections.namedtuple('IrcTuple',['index', 'row', 'col'])
XyzTuple = collections.namedtuple('XyzTuple',['x', 'y', 'z'])
origin_xyz = XyzTuple(*ct_mhd.GetOrigin())
vxSize_xyz = XyzTuple(*ct_mhd.GetSpacing())
direction_a = np.array(ct_mhd.GetDirection()).reshape(3, 3)
从CT扫描中取出⼀个结节
xyz2irc和irc2xyz在util.py中能找到源码;
def getRawCandidate(center_xyz, width_irc):
center_irc = util.xyz2irc(
center_xyz,
origin_xyz,
vxSize_xyz,
direction_a,
)
slice_list = []
for axis, center_val in enumerate(center_irc):
start_ndx = int(round(center_val - width_irc[axis]/2))
end_ndx = int(start_ndx + width_irc[axis])
assert center_val >= 0 and center_val < hu_a.shape[axis], repr([series_uid, center_xyz, origin_xyz, vxSize_xyz, center_irc, axis])
if start_ndx < 0:
# log.warning("Crop outside of CT array: {} {}, center:{} shape:{} width:{}".format(
# series_uid, center_xyz, center_irc, hu_a.shape, width_irc))
start_ndx = 0
end_ndx = int(width_irc[axis])
if end_ndx > hu_a.shape[axis]:
# log.warning("Crop outside of CT array: {} {}, center:{} shape:{} width:{}".format(
# series_uid, center_xyz, center_irc, hu_a.shape, width_irc))
end_ndx = hu_a.shape[axis]
start_ndx = int(hu_a.shape[axis] - width_irc[axis])
slice_list.append(slice(start_ndx, end_ndx))
ct_chunk = hu_a[tuple(slice_list)]
return ct_chunk, center_irc
a = getRawCandidate((-30,-30,-60),(10,10,10))
a[0].shape == (10,10,10),即 ct_chunk;
a[1] == IrcTuple(index=162, row=507, col=200),即 xyz2irc 后的 center_irc;
定义getCandidateInfoList
函数中设置了一些数据清洗的规则:
对于给定series_uid的每个候选条⽬,我们循环遍历之前为同⼀个series_uid收集的标注,看看2个坐标是否⾜够接近,如果⾜够接近则可以认为它们是相同的结节。如果是同⼀个结节,那太好了!现在我们有了这个结节的直径信息。如果没有找到匹配的结节,也没有关系,我们把结节的直径设置为0.
def getCandidateInfoList(requireOnDisk_bool=True):
# 构建一个存放当前所有uid的集合
mhd_list = glob.glob(path+'subset*/*.mhd')
presentOnDisk_set = {os.path.split(p)[-1][:-4] for p in mhd_list}
diameter_dict = {}
with open(path+'annotations.csv', "r") as f:
for row in list(csv.reader(f))[1:]:
series_uid = row[0]
annotationCenter_xyz = tuple([float(x) for x in row[1:4]])
annotationDiameter_mm = float(row[4])
diameter_dict.setdefault(series_uid, []).append(
(annotationCenter_xyz, annotationDiameter_mm)
)
candidateInfo_list = []
with open(path+'candidates.csv', "r") as f:
for row in list(csv.reader(f))[1:]:
series_uid = row[0]
if series_uid not in presentOnDisk_set and requireOnDisk_bool:
continue
isNodule_bool = bool(int(row[4]))
candidateCenter_xyz = tuple([float(x) for x in row[1:4]])
candidateDiameter_mm = 0.0
for annotation_tup in diameter_dict.get(series_uid, []):
annotationCenter_xyz, annotationDiameter_mm = annotation_tup
for i in range(3):
delta_mm = abs(candidateCenter_xyz[i] - annotationCenter_xyz[i])
if delta_mm > annotationDiameter_mm / 4:
# 将直径除以2得到半径,将半径除以2以要求2个结节中⼼点相对结节⼤⼩的距离不要太远。这将导致
# 边界框检查,⽽不是真正的距离检查
break
else:
candidateDiameter_mm = annotationDiameter_mm
break
candidateInfo_list.append(CandidateInfoTuple(
isNodule_bool,
candidateDiameter_mm,
series_uid,
candidateCenter_xyz,
))
candidateInfo_list.sort(reverse=True)
return candidateInfo_list
封装成类的完整程序:
前几步是将下列的class Ct拆成各部分单独解析,现在写成完整的一个类,导入LUNA16数据:
import torch
from torch.utils.data import Dataset
from logconf import logging
from util import XyzTuple, xyz2irc
log = logging.getLogger(__name__)
class Ct:
def __init__(self, series_uid):
mhd_path = glob.glob(
path+'subset*/{}.mhd'.format(series_uid)
)[0]
ct_mhd = sitk.ReadImage(mhd_path)
ct_a = np.array(sitk.GetArrayFromImage(ct_mhd), dtype=np.float32)
# 在做CT检查时,Hu是反映人体各组织的密度的指标
ct_a.clip(-1000, 1000, ct_a) # Hu的正常值为-1000~+1000
self.series_uid = series_uid
self.hu_a = ct_a
self.origin_xyz = XyzTuple(*ct_mhd.GetOrigin())
self.vxSize_xyz = XyzTuple(*ct_mhd.GetSpacing())
self.direction_a = np.array(ct_mhd.GetDirection()).reshape(3, 3)
def getRawCandidate(self, center_xyz, width_irc):
center_irc = xyz2irc(
center_xyz,
self.origin_xyz,
self.vxSize_xyz,
self.direction_a,
)
slice_list = []
for axis, center_val in enumerate(center_irc):
start_ndx = int(round(center_val - width_irc[axis]/2))
end_ndx = int(start_ndx + width_irc[axis])
assert center_val >= 0 and center_val < self.hu_a.shape[axis], repr([self.series_uid, center_xyz, self.origin_xyz, self.vxSize_xyz, center_irc, axis])
if start_ndx < 0:
# log.warning("Crop outside of CT array: {} {}, center:{} shape:{} width:{}".format(
# self.series_uid, center_xyz, center_irc, self.hu_a.shape, width_irc))
start_ndx = 0
end_ndx = int(width_irc[axis])
if end_ndx > self.hu_a.shape[axis]:
# log.warning("Crop outside of CT array: {} {}, center:{} shape:{} width:{}".format(
# self.series_uid, center_xyz, center_irc, self.hu_a.shape, width_irc))
end_ndx = self.hu_a.shape[axis]
start_ndx = int(self.hu_a.shape[axis] - width_irc[axis])
slice_list.append(slice(start_ndx, end_ndx))
ct_chunk = self.hu_a[tuple(slice_list)]
return ct_chunk, center_irc
# @functools.lru_cache(1, typed=True) #存入缓存,以免重复加载数据
def getCt(series_uid):
return Ct(series_uid)
# @raw_cache.memoize(typed=True)
def getCtRawCandidate(series_uid, center_xyz, width_irc):
ct = getCt(series_uid)
ct_chunk, center_irc = ct.getRawCandidate(center_xyz, width_irc)
return ct_chunk, center_irc
我们在这里通过指定每⼀组第10个样本作为验证集的成员来实现这⼀点,由val_stride参数指定;
我们还将接收一个isValSet_bool参数,并使用它来确定是否应该只保留训练数据、验证数据还是保留所有数据:
class LunaDataset(Dataset):
def __init__(self,
val_stride=0,
isValSet_bool=None,
series_uid=None,
):
self.candidateInfo_list = copy.copy(getCandidateInfoList())
# 复制返回值,这样缓存的副本就不会因为修改self.candidateInfo_list⽽受到影响
if series_uid:
self.candidateInfo_list = [
x for x in self.candidateInfo_list if x.series_uid == series_uid
]
if isValSet_bool:
assert val_stride > 0, val_stride
self.candidateInfo_list = self.candidateInfo_list[::val_stride]
assert self.candidateInfo_list
elif val_stride > 0:
del self.candidateInfo_list[::val_stride]
assert self.candidateInfo_list
log.info("{!r}: {} {} samples".format(
self,
len(self.candidateInfo_list),
"validation" if isValSet_bool else "training",
))
def __len__(self):
return len(self.candidateInfo_list)
def __getitem__(self, ndx):
candidateInfo_tup = self.candidateInfo_list[ndx]
width_irc = (32, 48, 48)
candidate_a, center_irc = getCtRawCandidate(
candidateInfo_tup.series_uid,
candidateInfo_tup.center_xyz,
width_irc,
)
candidate_t = torch.from_numpy(candidate_a)
candidate_t = candidate_t.to(torch.float32)
candidate_t = candidate_t.unsqueeze(0) # 将单通道的数据转换为多通道的数据,以满足某些模型的输入要求
# pos_t输出yes/no变成onehot型, [0,1]代表是结节,[1,0]代表不是结节
pos_t = torch.tensor([
not candidateInfo_tup.isNodule_bool, # 不是结节
candidateInfo_tup.isNodule_bool # 是结节
],
dtype=torch.long, # 变成长整型
)
return (
candidate_t,
pos_t,
candidateInfo_tup.series_uid,
torch.tensor(center_irc),
)
试打印 LunaDataset()[0] :分别返回输出
candidate_t,
pos_t,
candidateInfo_tup.series_uid,
torch.tensor(center_irc),
(tensor([[[[-732., -810., -863., ..., -848., -865., -890.],
[-767., -826., -855., ..., -908., -896., -879.],
[-807., -859., -848., ..., -898., -903., -897.],
...,
[-874., -869., -839., ..., -159., -121., -104.],
[-923., -930., -911., ..., -115., -104., -117.],
[-898., -898., -901., ..., -89., -93., -129.]],
[[-776., -779., -860., ..., -852., -875., -874.],
[-839., -834., -889., ..., -875., -894., -898.],
[-861., -857., -882., ..., -838., -865., -889.],
...,
[-862., -932., -917., ..., -111., -94., -101.],
[-896., -918., -898., ..., -16., 1., -41.],
[-875., -894., -881., ..., -55., -47., -83.]],
[[-854., -898., -884., ..., -886., -878., -857.],
[-856., -876., -865., ..., -869., -880., -884.],
[-841., -859., -881., ..., -857., -844., -832.],
...,
[-744., -880., -873., ..., -95., -55., -40.],
[-858., -895., -870., ..., -57., 4., 4.],
[-892., -897., -888., ..., -54., -23., -22.]],
...,
[[-901., -885., -877., ..., -11., 5., 20.],
[-909., -883., -881., ..., -14., -3., 29.],
[-925., -911., -895., ..., -35., -1., 31.],
...,
[ 373., 436., 473., ..., 17., 29., 26.],
[ 413., 439., 458., ..., -39., -20., -11.],
[ 334., 296., 302., ..., 7., 20., 36.]],
[[-911., -909., -894., ..., -16., -16., 11.],
[-935., -920., -903., ..., 12., 6., 20.],
[-936., -919., -886., ..., 6., 26., 33.],
...,
[ 0., 37., 49., ..., 26., 41., 32.],
[ 60., 36., 51., ..., -4., -6., -9.],
[ 213., 153., 114., ..., -31., -12., 1.]],
[[-949., -950., -956., ..., 11., 12., 23.],
[-948., -950., -941., ..., -6., -6., 16.],
[-924., -923., -904., ..., -40., -47., -9.],
...,
[ -77., -85., -98., ..., 48., 72., 73.],
[ -81., -112., -106., ..., 50., 72., 85.],
[ -55., -47., -56., ..., 86., 96., 96.]]]]),
tensor([0, 1]),
'1.3.6.1.4.1.14519.5.2.1.6279.6001.287966244644280690737019247886',
tensor([ 91, 360, 341]))
其中LunaDataset()[0][0].shape 为 torch.Size([1, 32, 48, 48]),1代表unsqueeze()添加的1维。