服务医学,基于目标检测模型实现细胞检测识别
在我以往的认知里面,显微镜下面的世界是很神奇的存在,可能只平时接触到的绝大多数是都是宏观的世界吧,看到微观世界里面各色各样的生物、细胞就会觉得很神奇,电子显微镜往往都是医生来操作观察的,对于采样、病理切片等任务比较繁重的时候,人工就会显得捉襟见肘了,这时候AI就可以提供帮助了,基于大量的医学数据进行学习拟合,得到的模型甚至能够超越领域专家,这些在很多主营医疗业务的公司里面都得到的实现,本文今天并不是谈很宏大的东西,就是做细胞数据集的目标检测,医学相关的数据感觉很有意思,后面有机会我也会多去做这个领域的项目,
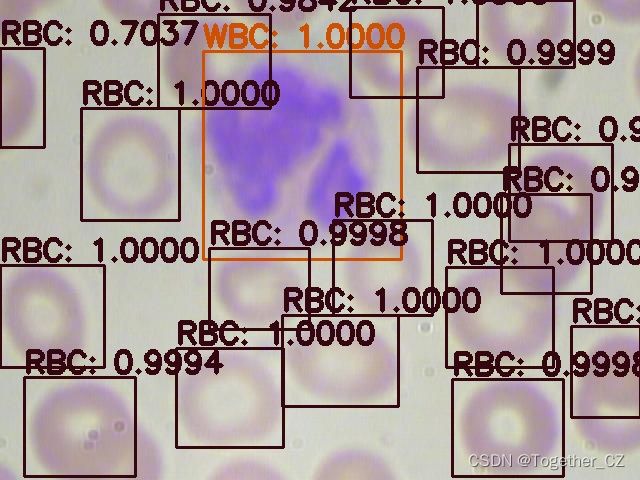
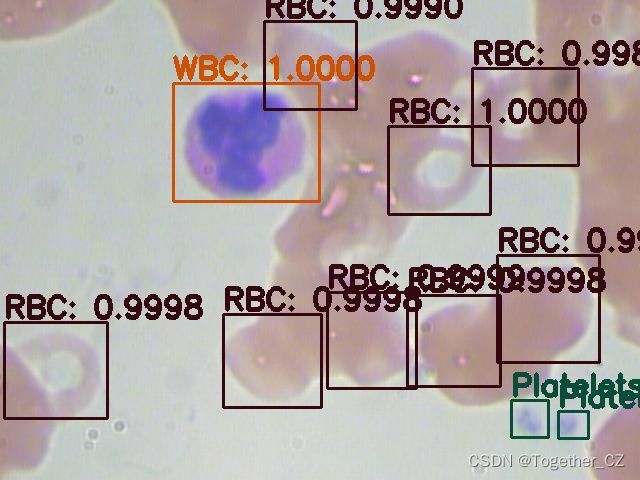
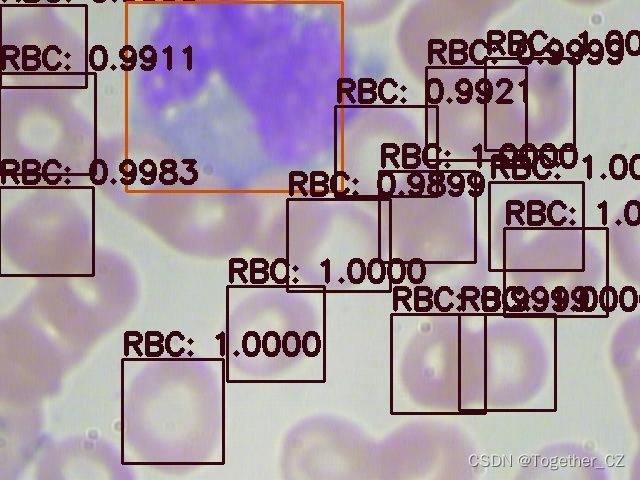
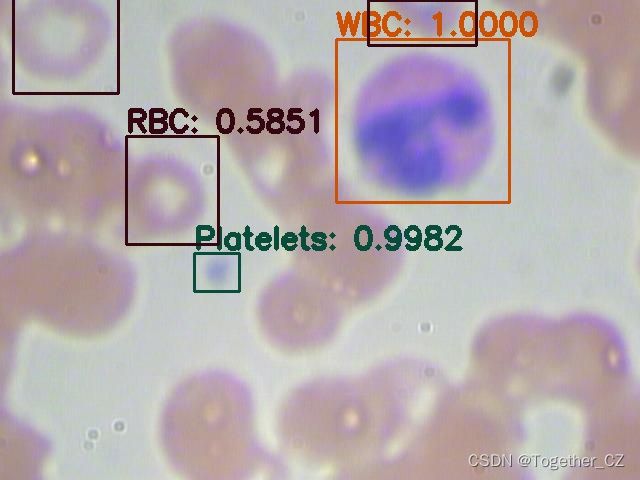
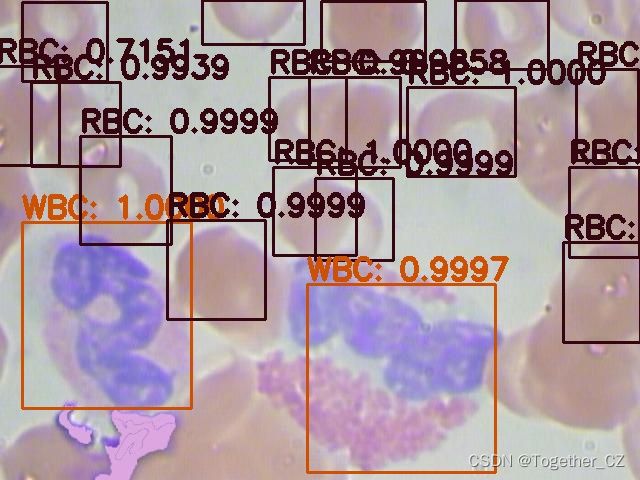
首先看下效果图:
这里面一共标注了三种细胞类型:
红细胞
血小板
白细胞英文表示分别为:
RBC
Platelets
WBC接下来看下数据集:
最近都在做一些轻量级的模型,这里模型选用的是MobileNetV2-YOLOv3-Lite的,详情如下所示:
[net]
# Training
batch=128
subdivisions=2
width=320
height=320
channels=3
momentum=0.9
decay=4e-5
angle=0
saturation = 1.5
exposure = 1.5
hue=.1
learning_rate=0.001
burn_in=1000
max_batches = 50200
policy=steps
steps=40000,45000
scales=.1,.1
[convolutional]
filters=32
size=3
stride=2
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=32
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=32
size=3
groups=32
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=16
size=1
stride=1
pad=1
batch_normalize=1
activation=linear
[convolutional]
filters=96
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=96
size=3
groups=96
stride=2
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=24
size=1
stride=1
pad=1
batch_normalize=1
activation=linear
[convolutional]
filters=144
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=144
size=3
groups=144
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=24
size=1
stride=1
pad=1
batch_normalize=1
activation=linear
[shortcut]
from=-4
activation=linear
[convolutional]
filters=144
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=144
size=3
groups=144
stride=2
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=32
size=1
stride=1
pad=1
batch_normalize=1
activation=linear
[convolutional]
filters=192
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=192
size=3
groups=192
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=32
size=1
stride=1
pad=1
batch_normalize=1
activation=linear
[shortcut]
from=-4
activation=linear
[convolutional]
filters=192
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=192
size=3
groups=192
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=32
size=1
stride=1
pad=1
batch_normalize=1
activation=linear
[shortcut]
from=-4
activation=linear
[convolutional]
filters=192
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=192
size=3
groups=192
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=64
size=1
stride=1
pad=1
batch_normalize=1
activation=linear
[convolutional]
filters=384
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=384
size=3
groups=384
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=64
size=1
stride=1
pad=1
batch_normalize=1
activation=linear
[shortcut]
from=-4
activation=linear
[convolutional]
filters=384
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=384
size=3
groups=384
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=64
size=1
stride=1
pad=1
batch_normalize=1
activation=linear
[shortcut]
from=-4
activation=linear
[convolutional]
filters=384
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=384
size=3
groups=384
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=64
size=1
stride=1
pad=1
batch_normalize=1
activation=linear
[shortcut]
from=-4
activation=linear
[convolutional]
filters=384
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=384
size=3
groups=384
stride=2
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=96
size=1
stride=1
pad=1
batch_normalize=1
activation=linear
[convolutional]
filters=576
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=576
size=3
groups=576
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=96
size=1
stride=1
pad=1
batch_normalize=1
activation=linear
[shortcut]
from=-4
activation=linear
[convolutional]
filters=576
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=576
size=3
groups=576
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=96
size=1
stride=1
pad=1
batch_normalize=1
activation=linear
[shortcut]
from=-4
activation=linear
[convolutional]
filters=576
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=576
size=3
groups=576
stride=2
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=160
size=1
stride=1
pad=1
batch_normalize=1
activation=linear
[convolutional]
filters=960
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=960
size=3
groups=960
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=160
size=1
stride=1
pad=1
batch_normalize=1
activation=linear
[shortcut]
from=-4
activation=linear
[convolutional]
filters=960
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=960
size=3
groups=960
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=160
size=1
stride=1
pad=1
batch_normalize=1
activation=linear
[shortcut]
from=-4
activation=linear
### SPP ###
[maxpool]
stride=1
size=3
[route]
layers=-2
[maxpool]
stride=1
size=5
[route]
layers=-4
[maxpool]
stride=1
size=9
[route]
layers=-1,-3,-5,-6
### End SPP ###
#################################
[convolutional]
filters=288
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=288
size=3
groups=288
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=96
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=384
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
size=1
stride=1
pad=1
filters=24
activation=linear
[yolo]
mask = 3,4,5
anchors = 26, 48, 67, 84, 72,175, 189,126, 137,236, 265,259
classes=3
num=6
jitter=.1
ignore_thresh = .5
truth_thresh = 1
random=1
#################
scale_x_y = 1.0
iou_thresh=0.213
cls_normalizer=1.0
iou_normalizer=0.07
iou_loss=ciou
nms_kind=greedynms
beta_nms=0.6
##################################
[route]
layers= 65
[upsample]
stride=2
[route]
layers=-1,48
#################################
[convolutional]
filters=80
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=288
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=288
size=3
groups=288
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=192
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
filters=288
size=1
stride=1
pad=1
batch_normalize=1
activation=relu
[convolutional]
size=1
stride=1
pad=1
filters=24
activation=linear
[yolo]
mask = 0,1,2
anchors = 26, 48, 67, 84, 72,175, 189,126, 137,236, 265,259
classes=3
num=6
jitter=.1
ignore_thresh = .5
truth_thresh = 1
random=1
#################
scale_x_y = 1.0
iou_thresh=0.213
cls_normalizer=1.0
iou_normalizer=0.07
iou_loss=ciou
nms_kind=greedynms
beta_nms=0.6
同样一共需要修改两处参数配置
【第一处】
【第二处】
主要就是修改里面classes和filters这两个关键字段即可:
classes就是你要检测的目标对象数量,这里我要检测的目标对象数量是:3
filters的数值计算公式为: filters=(classes+5)*,在这里就是 (3+5)*3=24
接下来编写cell.names和cell.data文件用于框架训练需要。
cell.names如下所示:
RBC
Platelets
WBCcell.data如下所示:
classes= 3
train = /home/objDet/cell/train.txt
valid = /home/objDet/cell/test.txt
names = /home/objDet/cell/x.names
backup = /home/objDet/cell/model/yolo基于Darknet框架可以一键启动训练,命令如下:
chmod +x darknet
nohup ./darknet detector train x.data MobileNetV2-YOLOv3-Lite.cfg >> yolo.out &入股对数据集格式有疑问的可以翻看前面的文章,前面已经详细介绍过了,所以后面的文章就不在赘述了。
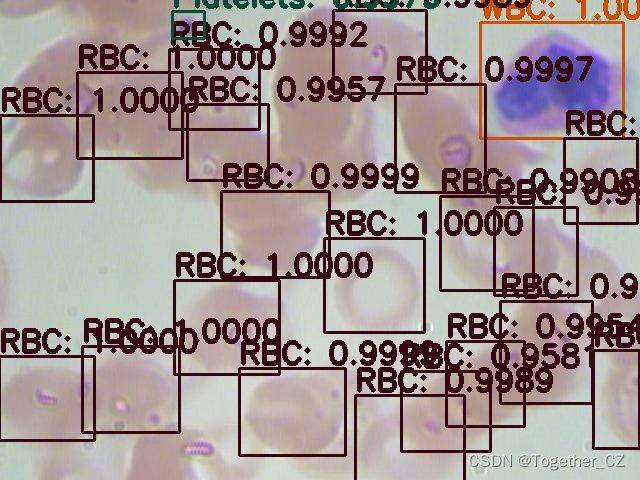
随机选择数据测试结果如下:
这里训练完成后, 我同样对整个训练过程的日志进行了可视化
我在上一篇文章《AI助力智能安检,基于目标检测模型实现X光安检图像智能检测分析》中,编写了原始日志的解析代码,如下:
def parseLogs(data="yolo.out"):
"""
解析原始日志
"""
with open(data) as f:
data_list=[one.strip() for one in f.readlines() if one.strip()]
print("data_list_length: ", len(data_list))
start=0
for i in range(len(data_list)):
if "hours left" in data_list[i]:
start=i
break
print("start: ", start)
s=start+1
res_list=[]
while s今天发现了可以有更简单的写法,代码如下:
def extractNeedLog(log_file,new_log_file,key_word):
"""
提取所需要的日志数据
"""
with open(log_file, 'r') as f:
with open(new_log_file, 'w') as train_log:
for line in f:
if key_word in line:
train_log.write(line)
f.close()
train_log.close()因为Darknet框架的训练日志是非常规则的文本形式,可以根据简单的关键词匹配即可完成所需信息的提取操作,不需要像我之前那样先分离了单个epoch的日志,之后再二次提取。
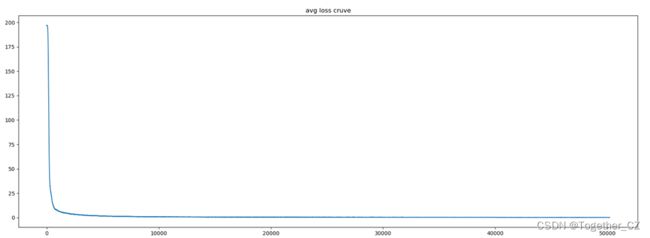
分离提取了loss日志和metric日志,这里对其进行可视化如下:
loss曲线
学习率曲线
时耗曲线
图像累积量曲线
其实这个没有必要可视化,简单分析就知道是一条直线了,因为每次选取的图片数是固定的,乘以总次数自然就是一条正比例直线了。
剩余时间曲线
这个是模型训练过程中不断打印输出的剩余训练时间的曲线,总体趋势降低就是对的了,中间有起伏跟服务器硬件有关系。
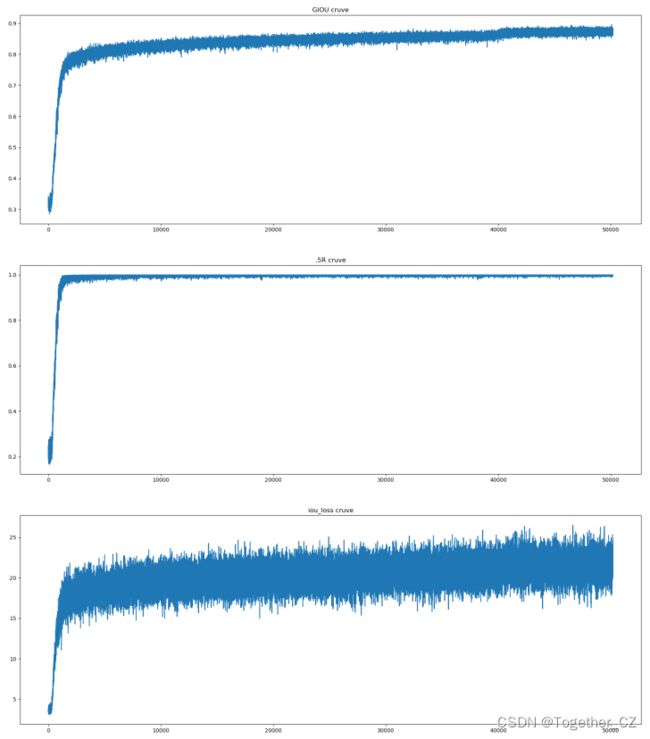
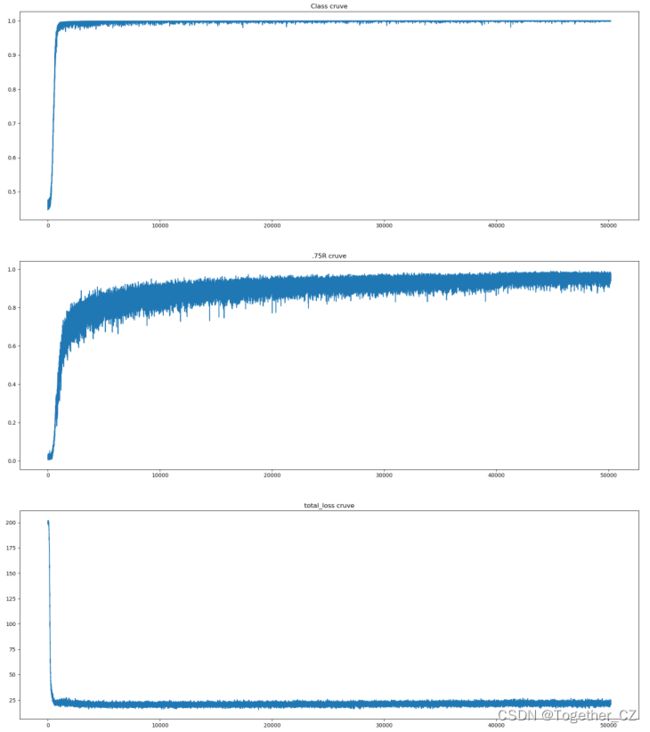
接下来是训练过程中的一些评估指标的可视化。
我是把觉得能可视化的都给可视化了,其实真实可能并不需要这么多,只需要关注几个重点需要关注的指标即可,比如:IOU,Obj、Class等等。