ggkegg | 用这个神包玩转kegg数据库吧!~(一)
1写在前面
好久没更了,实在是太忙了,值班真的是根本不不睡觉啊,一忙一整天,忙到怀疑人生。
最近看到比较的就是ggkegg包,感觉使用起来还是有一定难度的。
和大家分享一下使用教程吧,还有一些小坑。
2用到的包
rm(list = ls())
library(ggkegg)
library(ggfx)
library(ggraph)
library(igraph)
library(clusterProfiler)
library(dplyr)
library(tidygraph)
3小试牛刀
首先以eco00270为例,获取后转换路径和eco标识符,删除无连接节点并返回igraph对象。
g <- ggkegg(pid="eco00270",
convert_org = c("pathway","eco"),
delete_zero_degree = T,
return_igraph = T)
gg <- ggraph(g, layout="stress")
gg$data$type %>% unique()
gg + geom_edge_diagonal(
aes(color=subtype_name,
filter=type!="maplink"))+
geom_node_point(
aes(filter= !type%in%c("map","compound")),
fill=gg$data[!gg$data$type%in%c("map","compound"),]$bgcolor,
color="black",
shape=21, size=4
)+
geom_node_point(
aes(filter= !type%in%c("map","gene")),
fill=gg$data[!gg$data$type%in%c("map","gene"),]$bgcolor,
color="black",
shape=21, size=6
)+
geom_node_text(
aes(label=converted_name,
filter=type=="gene"),
repel=T,
bg.colour="white")+
theme_void()

4改变nodes颜色
颜狗必备技能,修改配色。
g <- pathway("ko00520")
V(g)$color_one <- colorRampPalette(RColorBrewer::brewer.pal(5,"Set1"))(length(V(g)))
V(g)$color_two <- colorRampPalette(RColorBrewer::brewer.pal(5,"Set2"))(length(V(g)))
ggraph(g, x=x, y=y) +
geom_node_rect(aes(xmin=xmin, xmax=x, fill=I(color_one)), alpha=0.5)+
geom_node_rect(aes(xmin=x, xmax=xmax, fill=I(color_two)), alpha=0.5)+
ggfx::with_outer_glow(geom_node_text(aes(label=name %>%
strsplit(":") %>%
sapply("[", 2) %>%
strsplit(" ") %>%
sapply("[", 1),
filter=type=="ortholog"),
size=2), colour="white", expand=1)

V(g)$color_one <- colorRampPalette(RColorBrewer::brewer.pal(5,"Set1"))(length(V(g)))
V(g)$color_two <- colorRampPalette(RColorBrewer::brewer.pal(5,"Set2"))(length(V(g)))
V(g)$color_three <- colorRampPalette(RColorBrewer::brewer.pal(5,"PuOr"))(length(V(g)))
V(g)$color_four <- colorRampPalette(RColorBrewer::brewer.pal(5,"Paired"))(length(V(g)))
V(g)$space <- V(g)$width/4
ggraph(g, x=x, y=y) +
geom_node_rect(aes(xmin=xmin, xmax=xmin+space, fill=I(color_one), filter=type=="ortholog"))+
geom_node_rect(aes(xmin=xmin+space, xmax=xmin+2*space, fill=I(color_two), filter=type=="ortholog"))+
geom_node_rect(aes(xmin=xmin+2*space, xmax=xmin+3*space, fill=I(color_three), filter=type=="ortholog"))+
geom_node_rect(aes(xmin=xmin+3*space, xmax=xmin+4*space, fill=I(color_four), filter=type=="ortholog"))+
ggfx::with_outer_glow(geom_node_text(aes(label=name %>%
strsplit(":") %>%
sapply("[", 2) %>%
strsplit(" ") %>%
sapply("[", 1),
filter=type=="ortholog"),
size=2), colour="white", expand=1)+
theme_void()

5Global maps展示
更宏观的展示结果。
pathway("ko01200") %>%
process_reaction() %>%
activate(nodes) %>%
mutate(x=NULL, y=NULL,
comp=convert_id("compound")) %>%
mutate(degree=centrality_degree(mode="all")) %>%
ggraph(layout="kk")+
geom_node_point(aes(color=degree,
filter=type=="compound"))+
geom_edge_parallel(
color="grey",
end_cap=circle(1,"mm"),
start_cap=circle(1,"mm"),
arrow=arrow(length=unit(1,"mm"),type="closed"))+
geom_node_text(aes(label=comp,filter=degree>15),
repel=TRUE, bg.colour="white")+
theme_graph()

6highlight指定nodes和edges
可以使用highligh_set_edges和highlight_set_nodes函数突出你想要突出的nodes和edges。
pathway("ko01230") %>%
process_line() %>%
activate(nodes) %>%
mutate(
compound=convert_id("compound"),
M00002=highlight_set_nodes(module("M00002")@reaction_components)) %>%
activate(edges) %>%
mutate(M00002=highlight_set_edges(module("M00002")@definition_components)) %>%
ggraph(x=x, y=y)+
geom_edge_link()+
with_outer_glow(geom_edge_link(aes(color=M00002, filter=M00002)),
colour="pink")+
geom_node_point(shape=21,aes(filter=type!="line"))+
with_outer_glow(geom_node_point(shape=21, aes(filter=M00002, color=M00002)),
colour="pink")+
geom_node_text(aes(label=compound, filter=M00002), repel=TRUE,
bg.colour="white", size=2)+
theme_void()

我们试着突出一下代谢相关pathway(ko01100),需要用到M00021的module。
g <- pathway("ko01100") %>%
process_line() %>%
highlight_module(module("M00021")) %>%
mutate(compound=convert_id("compound"))
g %>% ggraph(x=x, y=y) +
geom_node_point(size=1, aes(color=I(fgcolor),
filter=fgcolor!="none" & type!="line"))+
geom_edge_link(width=0.1, aes(color=I(fgcolor),
filter=type=="line"& fgcolor!="none"))+
with_outer_glow(
geom_edge_link(width=1,
aes(color=I(fgcolor),
filter=fgcolor!="none" & M00021)),
colour="red", expand=3
)+
with_outer_glow(
geom_node_point(size=2,
aes(color=I(fgcolor),
filter=fgcolor!="none" & M00021)),
colour="red", expand=3
)+
theme_void()

可以看到多个module涉及Cysteine和methionine的代谢,我们可以通过使用ggforce来高亮M00017。
list_of_modules <- c("M00021","M00338","M00609","M00017","M00034","M00035","M00368")
for (mm in list_of_modules) {
g <- g %>% highlight_module(module(mm))
}
ggraph(g,x=x,y=y,layout="manual") +
geom_edge_link0(width=0.5, color="grey")+
geom_edge_link(color="red",aes(filter=M00017|M00021|M00338|M00609|M00034|M00035|M00368))+
geom_node_point(size=2, color="red",aes(filter=M00017|M00021|M00338|M00609|M00034|M00035|M00368))+
ggforce::geom_mark_rect(aes(fill=M00017,
label=module("M00017")@name,
x=x, y=y,
group=M00017,
filter=M00017),
label.fill = "transparent",
label.fontsize = 10,
expand=unit(1,"mm"))+
theme_void()

可视化一下compounds,可以用geom_node_text,ggrepel, 和shadowtext。
Rg %>% ggraph(x=x, y=y) +
geom_node_point(size=1, aes(color=I(fgcolor),
filter=fgcolor!="none" & type!="line"))+
geom_edge_link(width=0.1, aes(color=I(fgcolor),
filter=type=="line"& fgcolor!="none"))+
with_outer_glow(
geom_edge_link(width=1,
aes(color=I(fgcolor),
filter=fgcolor!="none" & M00021)),
colour="red", expand=3
)+
with_outer_glow(
geom_node_point(size=2,
aes(color=I(fgcolor),
filter=fgcolor!="none" & M00021)),
colour="red", expand=3
)+
geom_node_text(aes(label=compound, filter=M00021),
repel=T, bg.colour="white", size=5)+
theme_void()
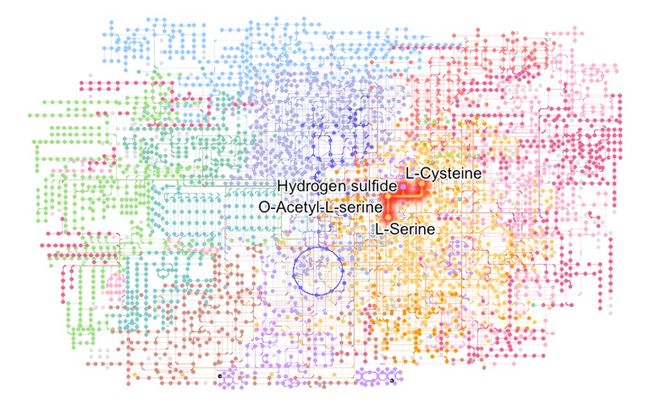
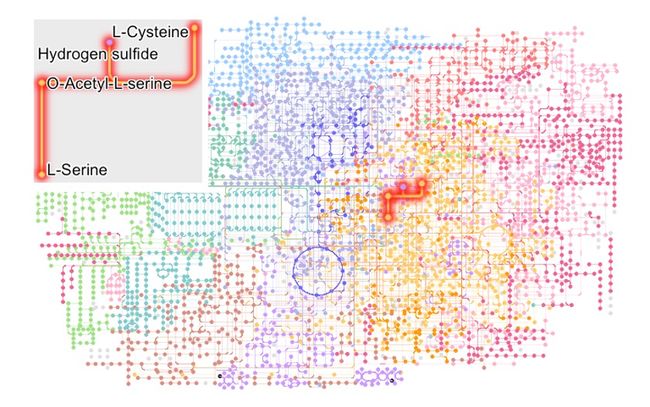
局部放大一下!~
annot <- g %>% ggraph(x=x, y=y)+
with_outer_glow(
geom_edge_link(width=1,
aes(color=I(fgcolor),
filter=fgcolor!="none" & M00021)),
colour="red", expand=3
)+
with_outer_glow(
geom_node_point(size=2,
aes(color=I(fgcolor),
filter=fgcolor!="none" & M00021)),
colour="red", expand=3
)+
geom_node_text(aes(label=compound, filter=M00021),
repel=TRUE, bg.colour="white", size=5)
g %>%
ggraph(x=x, y=y) +
geom_node_point(size=1, aes(color=I(fgcolor),
filter=fgcolor!="none" & type!="line"))+
geom_edge_link(width=0.1, aes(color=I(fgcolor),
filter=type=="line"& fgcolor!="none"))+
with_outer_glow(
geom_edge_link(width=1,
aes(color=I(fgcolor),
filter=fgcolor!="none" & M00021)),
colour="red", expand=3
)+
with_outer_glow(
geom_node_point(size=2,
aes(color=I(fgcolor),
filter=fgcolor!="none" & M00021)),
colour="red", expand=3
)+
annotation_custom(ggplotify::as.grob(annot),
ymin=-1500, ymax=0, xmin=0, xmax=1500)+
theme_void()
点个在看吧各位~ ✐.ɴɪᴄᴇ ᴅᴀʏ 〰
LASSO | 不来看看怎么美化你的LASSO结果吗!?
chatPDF | 别再自己读文献了!让chatGPT来帮你读吧!~
WGCNA | 值得你深入学习的生信分析方法!~
ComplexHeatmap | 颜狗写的高颜值热图代码!
ComplexHeatmap | 你的热图注释还挤在一起看不清吗!?
Google | 谷歌翻译崩了我们怎么办!?(附完美解决方案)
scRNA-seq | 吐血整理的单细胞入门教程
NetworkD3 | 让我们一起画个动态的桑基图吧~
RColorBrewer | 再多的配色也能轻松搞定!~
rms | 批量完成你的线性回归
CMplot | 完美复刻Nature上的曼哈顿图
Network | 高颜值动态网络可视化工具
boxjitter | 完美复刻Nature上的高颜值统计图
linkET | 完美解决ggcor安装失败方案(附教程)
......



本文由 mdnice 多平台发布