PE测序中read1与read2关系
PE测序中read1与read2关系
- 1、前言
- 2、插入片段与PE测序的3种关系
- 3、插入片段不测通时PE_reads间关系
-
- (1)fq中的reads
- (2)bam中的reads
- (3)IGV中的reads
- 参考文件
1、前言
涉及的基础概念部分,详见参考文件1;
插入片段介绍,详见参考文件2;
reads1、reads2即为对某个插入片段测序的结果。本文着重介绍1个插入片段的fastq=>bam的过程。为了加深理解,仿照参考文件3,拿自己数据复现一遍。
至于为什么有这篇文章,原因借用思考问题的熊的话:
很多东西就是这样,你以为的明白并不是真的明白,一年前的明白和一年后的明白也不是同一个明白。我这么说,不知道你能明白还是不明白
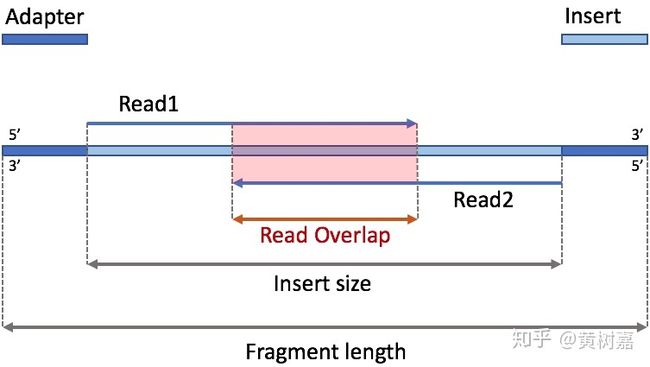
2、插入片段与PE测序的3种关系
数据量固定下,在插入片段略大于PE测序读长时(不测通),测序效过最好;
测通时最差,得到的reads数据都是被接头污染,需要舍弃掉,直接分析对结果有影响(SC序列比例高)。原因是,测序数据只会去掉5’端接头序列,reads的末端保留3’端的接头序列,比对时会认为是Softclip序列。
(1)不测通
(2)有重叠
(3)测通
3、插入片段不测通时PE_reads间关系
(1)fq中的reads
# read1
@A00812:653:HCCHGDSX5:1:1261:18285:34068 1:N:0:TTAGGTCG+AGTGACCT
GAGAGATCAGCTAGTGGGAAGGCAGCCTGGTCCCTGGTGTCAGGAAAATGCTGGCTGACCTAAAGCCACCTCCTTACTTTGCCTCCTTCTGCATGGTATTCTTTCTCTTCCGCACCCAGCAGTTTGGCCAGCCCAAAATCTGTGATCTTGA
+
FFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFF:FFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFF:FFFFFFFFFFFFFFFFFFFFFFFFFFFFF
# read2
@A00812:653:HCCHGDSX5:1:1261:18285:34068 2:N:0:TTAGGTCG+AGTGACCT
TTACCTTACTTGGAGGACCGTCGCTTGGTGCACCGCGACCTGGCAGCCAGGAACGTACTGGTGAAAACACCGCAGCATGTCAAGATCACAGATTTTGGGCTGGCCAAACTGCTGGGTGCGGACGAGAAAGAATACCATGCAGAAGGAGGCA
+
FFFFFFFFFFFFFFFFFFFFFFFF,FFFFFFFF:FFFFFFFFFFFFFFF,F::FFFFFFFF:FFFFFFFFFFFFFFF:FFFFFFF:FFFFFFF:FFFFFFFFFFFFFF:F:FFF,::FF:FF,FFFFFFFFFFF:FFFFFFFFFF,FFFFF
(2)bam中的reads
# bam中read1(原序列反向互补)
A00812:653:HCCHGDSX5:1:1261:18285:34068 83 chr7 55259494 60 141M = 55259425 -210
TCAAGATCACAGATTTTGGGCTGGCCAAACTGCTGGGTGCGGAAGAGAAAGAATACCATGCAGAAGGAGGCAAAGTAAGGAGGTGGCTTTAGGTCAGCCAGCATTTTCCTGACACCAGGGACCAGGCTGCCTTCCCACTAG
FFFFFFFFFFFFFFFFFFFFFFFFFFFFF:FFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFF:FFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFF
NM:i:0 MD:Z:141 MC:Z:141M AS:i:141 XS:i:0 RG:Z:EDT27243129B1D2L4
# bam中read2
A00812:653:HCCHGDSX5:1:1261:18285:34068 163 chr7 55259425 60 141M = 55259494 210
TGGAGGACCGTCGCTTGGTGCACCGCGACCTGGCAGCCAGGAACGTACTGGTGAAAACACCGCAGCATGTCAAGATCACAGATTTTGGGCTGGCCAAACTGCTGGGTGCGGAAGAGAAAGAATACCATGCAGAAGGAGGCA
FFFFFFFFFFFFFF,FFFFFFFF:FFFFFFFFFFFFFFF,F::FFFFFFFF:FFFFFFFFFFFFFFF:FFFFFFF:FFFFFFF:FFFFFFFFFFFFFF:F:FFF,::FF:FFFFFFFFFFFFFF:FFFFFFFFFF,FFFFF
NM:i:0 MD:Z:141 MC:Z:141M AS:i:141 XS:i:0 RG:Z:EDT27243129B1D2L4
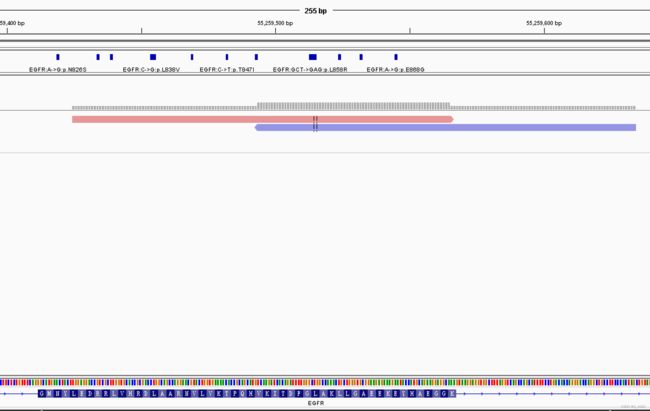
(3)IGV中的reads
红色的是reads2,从左到右,5’=>3’(正链);
蓝色的是reads1,原序列反向互补,从左到右,3’=>5’(负链);
read2序列
TGGAGGACCGTCGCTTGGTGCACCGCGACCTGGCAGCCAGGAACGTACTGGTGAAAACACCGCAGCATGTCAAGATCACAGATTTTGGGCTGGCCAAACTGCTGGGTGCGGAAGAGAAAGAATACCATGCAGAAGGAGGCA
read1反向互补序列
TCAAGATCACAGATTTTGGGCTGGCCAAACTGCTGGGTGCGGAAGAGAAAGAATACCATGCAGAAGGAGGCAAAGTAAGGAGGTGGCTTTAGGTCAGCCAGCATTTTCCTGACACCAGGGACCAGGCTGCCTTCCCACTAGCTGATCTCTC
ref序列
TGGAGGACCGTCGCTTGGTGCACCGCGACCTGGCAGCCAGGAACGTACTGGTGAAAACACCGCAGCATGTCAAGATCACAGATTTTGGGCTGGCCAAACTGCTGGGTGCGGAAGAGAAAGAATACCATGCAGAAGGAGGCAAAGTAAGGAGGTGGCTTTAGGTCAGCCAGCATTTTCCTGACACCAGGGACCAGGCTGCCTTCCCACTAGCTGTATTGTT
结论:
(1)PE测序的read1和read2是两条互补链;
(2)insertsize中方向相对的两条序列,比对到单链的参考基因组之前会先将其中一条read转义,然后进行比对;BAM文件的read1、read2中有一条是反向互补的。
(3)从UCSC、NCBI或Ensembl下载的参考基因组都是正链碱基序列(5’=>3’)
参考文件
1、生信学习笔记:fastp质控处理生成的report结果解读_fastp结果解读_twocanis的博客-CSDN博客
2、一篇文章说清楚什么是“插入片段”? - 知乎 (zhihu.com)
3、双端测序中read1和read2的关系 - 简书 (jianshu.com)