source localization by MNE
source localization by MNE
https://www.nmr.mgh.harvard.edu/mne/0.14/auto_tutorials/plot_mne_dspm_source_localization.html
// An highlighted block
# https://www.nmr.mgh.harvard.edu/mne/0.14/auto_tutorials/plot_mne_dspm_source_localization.html
import numpy as np
import matplotlib.pyplot as plt
import mne
from mne.datasets import sample
from mne.minimum_norm import (make_inverse_operator, apply_inverse,
write_inverse_operator)
# data_path = sample.data_path()
# raw_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
raw_fname ='E:\\data-c\\mne_data\\MNE-sample-data\\MEG\\sample\\sample_audvis_filt-0-40_raw.fif'
raw = mne.io.read_raw_fif(raw_fname,preload=True)
raw.set_eeg_reference() # set EEG average reference
events = mne.find_events(raw, stim_channel='STI 014')
event_id = dict(aud_r=1) # event trigger and conditions
tmin = -0.2 # start of each epoch (200ms before the trigger)
tmax = 0.5 # end of each epoch (500ms after the trigger)
raw.info['bads'] = ['MEG 2443', 'EEG 053']
picks = mne.pick_types(raw.info, meg=True, eeg=False, eog=True,
exclude='bads')
baseline = (None, 0) # means from the first instant to t = 0
reject = dict(grad=4000e-13, mag=4e-12, eog=150e-6)
epochs = mne.Epochs(raw, events, event_id, tmin, tmax, proj=True, picks=picks,
baseline=baseline, reject=reject)
noise_cov = mne.compute_covariance(
epochs, tmax=0., method=['shrunk', 'empirical'])
fig_cov, fig_spectra = mne.viz.plot_cov(noise_cov, raw.info)
evoked = epochs.average()
evoked.plot()
evoked.plot_topomap(times=np.linspace(0.05, 0.15, 5), ch_type='mag')
# Show whitening
evoked.plot_white(noise_cov)
# Read the forward solution and compute the inverse operator
data_path='E:\\data-c\\mne_data\\MNE-sample-data'
fname_fwd = data_path + '\\MEG\\sample\\sample_audvis-meg-oct-6-fwd.fif'
fwd = mne.read_forward_solution(fname_fwd)
# Restrict forward solution as necessary for MEG
fwd = mne.pick_types_forward(fwd, meg=True, eeg=False)
# make an MEG inverse operator
info = evoked.info
inverse_operator = make_inverse_operator(info, fwd, noise_cov,
loose=0.2, depth=0.8)
write_inverse_operator('sample_audvis-meg-oct-6-inv.fif',
inverse_operator,overwrite=True)
method = "dSPM"
snr = 3.
lambda2 = 1. / snr ** 2
stc = apply_inverse(evoked, inverse_operator, lambda2,
method=method, pick_ori=None)
# del fwd, inverse_operator, epochs # to save memory
plt.plot(1e3 * stc.times, stc.data[::100, :].T)
plt.xlabel('time (ms)')
plt.ylabel('%s value' % method)
plt.show()
#--------------------右脑-----------------------------------------------
# vertno_max, time_max = stc.get_peak(hemi='rh')
#
# subjects_dir = data_path + '/subjects'
# brain = stc.plot(surface='inflated', hemi='rh', subjects_dir=subjects_dir, initial_time=time_max,
# clim=dict(kind='value', lims=[8, 12, 15]),
# time_unit='s')
# brain.add_foci(vertno_max, coords_as_verts=True, hemi='rh', color='blue',
# scale_factor=0.6)
# brain.show_view('lateral')
#
# print('--------------------------------------')
#
# fs_vertices = [np.arange(10242)] * 2
# morph_mat = mne.compute_morph_matrix('sample', 'fsaverage', stc.vertices,
# fs_vertices, smooth=None,
# subjects_dir=subjects_dir)
# stc_fsaverage = stc.morph_precomputed('fsaverage', fs_vertices, morph_mat)
# brain_fsaverage = stc_fsaverage.plot(surface='inflated', hemi='rh',
# subjects_dir=subjects_dir,
# clim=dict(kind='value', lims=[8, 12, 15]),
# initial_time=time_max, time_unit='s')
# brain_fsaverage.show_view('lateral')
#--------------------------左脑-----------------------------------------------------
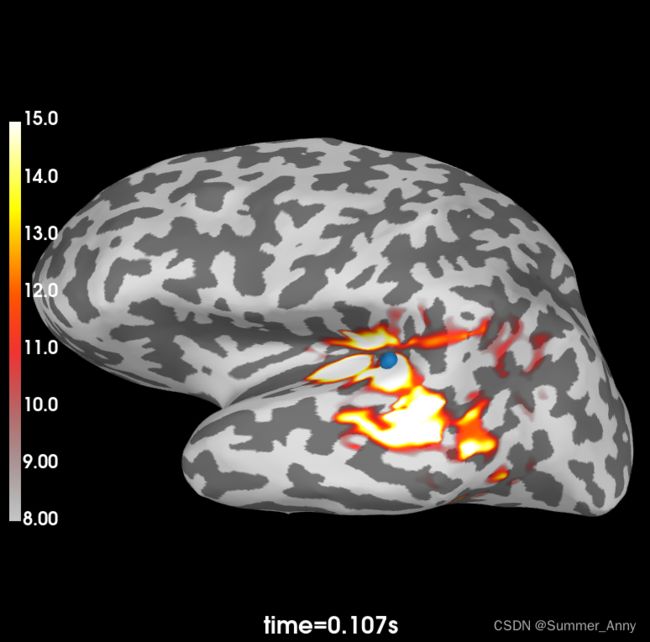
vertno_max, time_max = stc.get_peak(hemi='lh')
subjects_dir = data_path + '/subjects'
brain = stc.plot(surface='inflated', hemi='lh', subjects_dir=subjects_dir, initial_time=time_max,
clim=dict(kind='value', lims=[8, 12, 15]),
time_unit='s')
brain.add_foci(vertno_max, coords_as_verts=True, hemi='lh', color='blue',
scale_factor=0.6)
brain.show_view('lateral')
print('--------------------------------')
#
# fs_vertices = [np.arange(10242)] * 2
# morph_mat = mne.compute_morph_matrix('sample', 'fsaverage', stc.vertices,
# fs_vertices, smooth=None,
# subjects_dir=subjects_dir)
# stc_fsaverage = stc.morph_precomputed('fsaverage', fs_vertices, morph_mat)
# brain_fsaverage = stc_fsaverage.plot(surface='inflated', hemi='lh',
# subjects_dir=subjects_dir,
# clim=dict(kind='value', lims=[8, 12, 15]),
# initial_time=time_max, time_unit='s')
# brain_fsaverage.show_view('lateral')