- 领域模型、MDD\DDD\TDD概念
lemon_lmlmlmlm
java
此篇是个人笔记整理,知识来源:领域模型-CSDN博客、什么是MDD,DDD,TDD?-CSDN博客一、领域模型定义:领域模型是对领域内的概念类或现实世界中对象的可视化表示。又称概念模型、领域对象模型、分析对象模型。它专注于分析问题领域本身,发掘重要的业务领域概念,并建立业务领域概念之间的关系。领域模型(DomainModel),是完成从需求分析到面向对象设计的一座桥梁,领域模型是指对需求所涉及的领
- TwoSampleMR 报错解决:Error in r$status_code : $ operator is invalid for atomic vectors
生信碱移
r语言
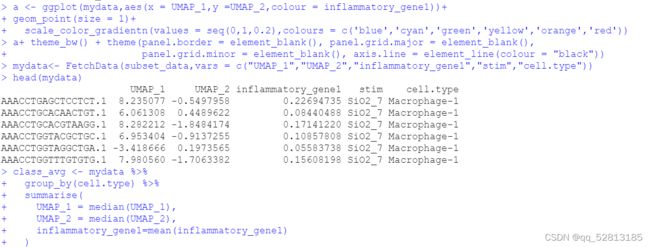
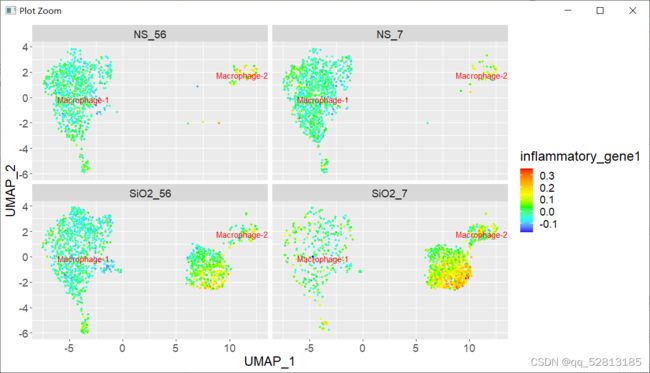
运行以下代码的时候报错:library(TwoSampleMR)ao<-available_outcomes()简单看了下available_outcomes()函数的源码,不是TwoSampleMR的问题,应该是ieugwasr::gwasinfo函数的问题:去ieugwasr包github的issue看了看,发现作者5天前就解决了这个问题:好吧,解决方案就是重新安装一下ieugwasr包#删除
- R 调用 python
weixin_33971205
pythonjsonjava
上一篇说了python使用rpy2调用R,这里介绍R如何调用python。R的强项在于统计方面,尤其是专业的统计分析,统计检验以及作图功能十分强大,但是在通用性方面,就远不如Python了,比如python可以做web,可以开发GUI,可以爬虫,甚至可以开发游戏,这些R其实也不是完全不行,但是在易用性方面实在是难以与Python相匹敌。所以如果要是能将R与Python相结合,充分发挥二者的优势,那
- AI会不会是一个大泡沫
人机与认知实验室
人工智能
深度求索(DeepSeek)引发的震撼,还在继续进行中。刚看了一下数字:美股盘前,英伟达跌超11%,博通跌超11%,台积电、Arm跌超10%,同时,在日本股市,英伟达的主要供应商AdvantestCorp的股价暴跌8.6%。这些科技股,真是跌惨了。这整件事,就像魔幻一下。1月20日,深度求索正式发布推理大模型DeepSeek-R1。推出后不久,R1就凭借其开源的性质、大幅下降的售价和训练成本获得了
- Java基础入门3《IDEA使用与数据类型详解》
圆圆Alice
Alice的Java学习笔记intellijideajava
Hi,我又出现了,是Alice,今天我学习的是,刚刚入门,肯定是先搞定各种开发工具,还一些理解性的内容,视频来源于‘蓝桥研究院’.....本节笔记重点提要:本节笔记重点提要:1.IntellijIdea下载与安装2.IntellijIdea使用3.使用IntellijIdea打印一个界面(案列)4.变量5.Java的数据类型、数据范围、符号的意义——————————————————————————
- Docker最新安装教程基于centos7.9
Acerola-
菜鸡上路docker
1.基本准备Docker要求内核版本3.10以上,请务必注意,安装教程基于centos7.9,内核版本3.10.0-1160.11.1.el7.x86_64#查看内核版本命令uname-r2.安装以下步骤依次进行#安装环境依赖yuminstall-yyum-utils#设置下载Docker的镜像源yum-config-manager\--add-repo\https://zfzbet67.mirr
- miniconda中成功安装gdal库
跳动的弦
遥感学习python
通过网上浏览相关帖子,未找到在miniconda中安装gdal库的相关内容,本人在本次安装成功之前已经历过两次失败,故在这里记录本次成功安装的细节,希望对有相似需求的朋友有所帮助。介于本人在这一方面也是初学者,有不正确的地方还请各位好友不吝指教。安装之前先下载安装RedistributingVisualC++Files,配置好VisualC++环境LatestsupportedVisualC++R
- k8s(kubernetes)常见故障处理总结——详细文档
运维实战课程
docker和k8s学习文档kubernetesdocker容器
k8s(kubernetes)常见故障处理总结——详细文档本文涵盖了k8s运维过程中大部分常见的故障,提供相应的排查思路,笔记详细,仅供参考本人会经常更新运维相关技术文档,如有兴趣,可以关注我博客,欢迎互动分享k8s其他相关详细资料下载地址:kubeadm初始化高可用k8s1.20.4集群网盘地址:https://url28.ctfile.com/f/37115828-599516373-25f4
- [论文笔记] llama-factory 微调qwen2.5、llama3踩坑
心心喵
论文笔记深度学习人工智能
一、bug1、pre-tokenize的时候,会OOM解决:在yaml文件中添加streaming参数#tokenizestreaming:Truemax_steps:10000https://github.com/hiyouga/LLaMA-Factory/blob/3a023bca2a502810a436cfba7708df164754ea62/src/llamafactory/hparams
- ogre 学习笔记 - Day 1
頖╃縌 ①oO%
#ogre学习笔记游戏引擎
ogre学习笔记-Day1OGRE:Object-OrientedGraphicsRenderingEngine从名称可以得出,OGRE是一个渲染引擎下载地址https://www.ogre3d.org/最新版ogre-13.1.0编译工具cmake-gui,vs2019,vscodeConfigureconfigure时发现ogre会自动从github下载/编译依赖项,github速度有可能很慢
- Nginx 学习笔记
韩某-
nginx学习笔记
目录一、引言二、Nginx概述三、Nginx的作用(一)正向代理(二)反向代理(三)负载均衡策略(四)动静分离四、Nginx安装五、Nginx的常用命令六、Nginx实战及总结一、引言在项目发展初期,并发量和用户量较少时,简单地将一个jar包部署到服务器tomcat上即可满足需求。然而,随着用户数量的不断增长以及并发量的持续增大,单台服务器容易面临性能瓶颈,出现“红温”现象。此时,为了提升系统的处
- 第一章:Reac入门 与 第二章:React面向组件编程
代码界小菜鸟
reactreact
目录一、jsx语法规则二、React中定义组件1.函数式组件:2.类式组件:*有关类复习的知识点前往React知识铺垫查看https://blog.csdn.net/m0_61927991/article/details/126591057?csdn_share_tail=%7B%22type%22%3A%22blog%22%2C%22rType%22%3A%22article%22%2C%22r
- Selenium学习笔记--Webdriver API 2--常用方法
jiang_guo
自动化测试笔记selenium
Webdriver重用方法浏览器控制控制浏览器窗口大小控制浏览器后退、前进切换标签页(窗口切换)switch方法获取url使用get方法模拟浏览器刷新关闭浏览器常用方法clearsend_keysclicksubmitsizetextget_attributeis_displayedtitlecurrent_url鼠标操作键盘操作元素等待显示等待隐式等待切换iframe单表单切换嵌套表单切换平行表
- 2021 React面试题知识点很全面
尘之微识
react.js面试
一,什么是虚拟DOM?虚拟DOM(VDOM)它是真实DOM的内存表示,一种编程概念,一种模式。它会和真实的DOM同步,比如通过ReactDOM这种库,这个同步的过程叫做调和(reconcilation)。虚拟DOM更多是一种模式,不是一种特定的技术。二,类组件和函数组件之间有什么区别?类组件(Classcomponents)无论是使用函数或是类来声明一个组件,它决不能修改它自己的props。所有R
- GSEA - Gene set enrichment analysis 基因集富集 | ORA - Over-Representation Analysis 分析原理与应用...
weixin_30294709
python数据库人工智能
R批量做GSEA分析还没有官方的包,但是clusterprofiler可以做,它调用了最新的gfsea包。GeneSetTestingforRNA-seq-fgsea教程RNA-seq是利器,大部分做实验的老板手下都有大量转录组数据,所以RNA-seq的分析需求应该是很大的(大部分的生信从业人员应该都差不多要沾边吧)。普通的转录组套路并不多,差异表达基因、富集分析、WGCNAnetwork以及一些
- windows环境下anaconda/miniconda 在gitbash的zsh中使用conda activate
大熊SsSsSs
windowsconda
这个问题折磨了一天防止以后再出现此问题忘记解决方法记录一下问题描述核心原因就是\r,\n,\r\n的区别可能会导致:(eval):11:parseerrornear^M解决方案解决方案如下:在~/.zshrc中添加下面这个(记得修改conda.exe的路径)#>>>condainitialize>>>#!!Contentswithinthisblockaremanagedby'condainit'
- Python下3种文字识别工具的源码和效果比较
eybk
python开发语言
1.pytesseractimportpytesseractfromPILimportImageim=Image.open(r'C:/Users/YBK/Pictures/35005.jpg')string=pytesseract.image_to_string(im,lang='chi_sim')print(string)2.paddleocrfrompaddleocrimportPaddleO
- 基于深度学习的遥感目标检测系统:UI界面、R-CNN模型与数据集准备
2025年数学建模美赛
R-CNN检测系统人工智能深度学习r语言cnnpythonui目标检测
一、引言遥感图像中的目标检测在很多领域,如环境监测、土地利用、城市规划、农业资源监测等方面有着广泛应用。遥感图像具有高分辨率和丰富的空间信息,但同时也带来了目标检测中的许多挑战,特别是在目标尺度变化、遮挡和复杂背景的情况下。因此,采用深度学习技术,尤其是卷积神经网络(CNN)和区域卷积神经网络(R-CNN),在遥感图像目标检测中取得了显著的成果。本文将详细介绍基于深度学习的遥感目标检测系统,使用R
- DeepSeek R1:中国AI黑马的崛起与挑战
码事漫谈
AI人工智能
文章目录技术突破:从零开始的推理能力进化DeepSeekR1-Zero:纯RL训练的“自我觉醒”DeepSeekR1:冷启动与多阶段训练的平衡之道实验验证:推理能力的全方位跃升基准测试:超越顶尖闭源模型蒸馏技术:小模型的逆袭行业启示:AGI之路的新范式纯RL训练的价值与挑战蒸馏技术的普惠意义开源生态的推动力未来展望:从推理到通用智能结语在人工智能领域,大型语言模型(LLMs)正以迅猛之势重塑我们的
- Shiro框架源码学习笔记
a88729845
shiro
文章目录介绍认证术语如何使用Shiro的认证1.手机认证主体和凭据2.提交认证主体和凭据到认证系统3.允许访问,重新认证,或阻止访问"RememberMeSupport"RememberedvsAuthenticated登出授权授权三要素权限权限的粒度角色隐式的角色显式的角色(推荐)用户Shiro如何执行授权编程式授权角色检查权限检查实现`Permission`接口的方式使用`String`表示一
- 【MySQL】性能测试方法
nickli528
mysql数据库
本页主要介绍关系数据库MySQL版的性能测试方法。测试环境测试使用ECS和RDS实例均在同一地域、同一可用区。测试使用ECS和RDS实例在同一VPC内。测试使用ECS信息:规格为s6.4xlarge.2镜像为CentOS7.9测试使用RDS信息:规格为S2/S7系列下各规格存储类型为超高IO数据盘400G参数模板为RDS默认参数模板说明1.ECS规格较高是为了保证测试时性能瓶颈不在ECS端。2.R
- 五、BLE Demo目录详解及演示
才才爱烤火
BLE入门到精通(基于TIcc26x2)linuxc语言mcustm32物联网嵌入式硬件iot
1、导入、编译、烧录打开之前安装的CCS,如下点击导入工程选择如下工程:simplelink_cc13xx_cc26xx_sdk_7_10_01_24\examples\rtos\CC26X2R1_LAUNCHXL\ble5stack\simple_peripheral如图常用的三个按钮2、BLE使用演示咱们详细介绍下目录:介绍完成后连接开发板点击下载,下载成功后打开串口工具,重新上电就可以看到我
- 2025年1月27日人工智能与科技新闻:DeepSeek震撼全球科技市场
海棠AI实验室
AI瞭望塔人工智能科技
中国AI初创公司DeepSeek震撼全球科技市场DeepSeekR1模型的突破性发展在科技迅猛发展的时代,中国人工智能初创公司DeepSeek推出的R1模型,以其卓越的技术能力,在全球AI领域掀起一场变革。这不仅是技术层面的突破,更对全球科技市场产生深远影响。技术创新与突破全新语言处理架构:提升AI理解和生成能力超越传统模型的认知能力:实现更精准的自然语言推理多模态交互:支持文本、图像、语音等多维
- 若依路由配置教程
yqj234
java前端服务器
1.路由配置文件2.配置内容介绍{path:"/tool/gen-edit",component:Layout,//在路由下,引用组件的名称,在页面中包括这个组件的内容(页面框架内容)hidden:true,//此页面的内容,在左边的菜单中不用显示。children:[{path:"index",//子路径,拼接在父路径后面的内容(/tool/gen-edit/index)component:(r
- Maui学习笔记-身份认证和授权案例
Mr.L70517
Maui学习笔记学习笔记iosc#http
在深入研究身份验证和授权时,可能会遇到很多术语。我们来简单介绍一下。Authentication,简单来讲时认证、验证身份检查用户名和密码,更高级方法设计到指纹、扫描、人脸识别或2FA认证。Authorization,授权,一旦通过身份认证,系统就可以决定当前用户是否有访问某些信息或执行一些操作的授权。OpenAuthorization(OAuth),开放授权,它允许第三方用户访问你的程序,而无需
- 链表的基础知识
erchazhan
链表网络数据结构
在大一学习链表的过程中,感觉有许多没有学过的知识,这篇文章,算是我的第一篇学习笔记,可以在后续学习中回顾,有不对的情况可以提出,谢谢大家的建议。#pragmaonce#include#include//#include"SList.h"typedefintSLDateType;//voidSListPrint(SListNode*phead);定义结构体typedefstructNode{SLDa
- 图文详解 MapReduce on YARN
Shockang
大数据技术体系大数据mapreduceyarn
前言本文隶属于专栏《1000个问题搞定大数据技术体系》,该专栏为笔者原创,引用请注明来源,不足和错误之处请在评论区帮忙指出,谢谢!本专栏目录结构和参考文献请见1000个问题搞定大数据技术体系正文权威版本——《Hadoop权威指南第3版》1.作业提交MRrunJob从RM获取新的作业ID作业客户端检査作业的输出说明,计算输入分片并将作业资源(包括作业JAR、配置和分片信息)复制到HDFS。通过调用R
- 【STL笔记】字符串
Flower#
算法学习笔记c++笔记
字符串下标从0开始,常规用法不再赘述,持续更新中…1.substr(pos,len):返回从位置pos开始,长度为len的子串。(len默认为npos)std::stringstr="Hello,World!";std::stringsub1=str.substr(7,5);//提取从索引7开始,长度为5的子串std::stringsub2=str.substr(7);//提取从索引7开始直到字符
- [补题]G2. Ruler (hard version)
Flower#
题解/补题c++算法c语言
G2.Ruler(hardversion)思路:hard和easy版的区别就是从10次查询变成7次。如果是10次,那么可以每次查询一个正方形用二分的方法容易的做出来。如果是7次,可以用类似于三分的方法,查询l×rl\timesrl×r的矩形的结果q:若q=lm∗rq=lm*rq=lm∗r则x>rmx>rmx>rm若q=lm∗(rm+1)q=lm*(rm+1)q=lm∗(rm+1)则lm=lmx>=
- {每日一道算法题}
zzh666ya
算法算法javapythonc++c语言
423.从英文中重建数字难度中等题目描述:给你一个字符串s,其中包含字母顺序打乱的用英文单词表示的若干数字(0-9)。按升序返回原始的数字提示:1<=s.length<=105s[i]为["e","g","f","i","h","o","n","s","r","u","t","w","v","x","z"]这些字符之一示例1:输入:s="owoztneoer"输出:"012"示例2:输入:s="f
- Enum用法
不懂事的小屁孩
enum
以前的时候知道enum,但是真心不怎么用,在实际开发中,经常会用到以下代码:
protected final static String XJ = "XJ";
protected final static String YHK = "YHK";
protected final static String PQ = "PQ";
- 【Spark九十七】RDD API之aggregateByKey
bit1129
spark
1. aggregateByKey的运行机制
/**
* Aggregate the values of each key, using given combine functions and a neutral "zero value".
* This function can return a different result type
- hive创建表是报错: Specified key was too long; max key length is 767 bytes
daizj
hive
今天在hive客户端创建表时报错,具体操作如下
hive> create table test2(id string);
FAILED: Execution Error, return code 1 from org.apache.hadoop.hive.ql.exec.DDLTask. MetaException(message:javax.jdo.JDODataSto
- Map 与 JavaBean之间的转换
周凡杨
java自省转换反射
最近项目里需要一个工具类,它的功能是传入一个Map后可以返回一个JavaBean对象。很喜欢写这样的Java服务,首先我想到的是要通过Java 的反射去实现匿名类的方法调用,这样才可以把Map里的值set 到JavaBean里。其实这里用Java的自省会更方便,下面两个方法就是一个通过反射,一个通过自省来实现本功能。
1:JavaBean类
1 &nb
- java连接ftp下载
g21121
java
有的时候需要用到java连接ftp服务器下载,上传一些操作,下面写了一个小例子。
/** ftp服务器地址 */
private String ftpHost;
/** ftp服务器用户名 */
private String ftpName;
/** ftp服务器密码 */
private String ftpPass;
/** ftp根目录 */
private String f
- web报表工具FineReport使用中遇到的常见报错及解决办法(二)
老A不折腾
finereportweb报表java报表总结
抛砖引玉,希望大家能把自己整理的问题及解决方法晾出来,Mark一下,利人利己。
出现问题先搜一下文档上有没有,再看看度娘有没有,再看看论坛有没有。有报错要看日志。下面简单罗列下常见的问题,大多文档上都有提到的。
1、没有返回数据集:
在存储过程中的操作语句之前加上set nocount on 或者在数据集exec调用存储过程的前面加上这句。当S
- linux 系统cpu 内存等信息查看
墙头上一根草
cpu内存liunx
1 查看CPU
1.1 查看CPU个数
# cat /proc/cpuinfo | grep "physical id" | uniq | wc -l
2
**uniq命令:删除重复行;wc –l命令:统计行数**
1.2 查看CPU核数
# cat /proc/cpuinfo | grep "cpu cores" | u
- Spring中的AOP
aijuans
springAOP
Spring中的AOP
Written by Tony Jiang @ 2012-1-18 (转)何为AOP
AOP,面向切面编程。
在不改动代码的前提下,灵活的在现有代码的执行顺序前后,添加进新规机能。
来一个简单的Sample:
目标类:
[java]
view plain
copy
print
?
package&nb
- placeholder(HTML 5) IE 兼容插件
alxw4616
JavaScriptjquery jQuery插件
placeholder 这个属性被越来越频繁的使用.
但为做HTML 5 特性IE没能实现这东西.
以下的jQuery插件就是用来在IE上实现该属性的.
/**
* [placeholder(HTML 5) IE 实现.IE9以下通过测试.]
* v 1.0 by oTwo 2014年7月31日 11:45:29
*/
$.fn.placeholder = function
- Object类,值域,泛型等总结(适合有基础的人看)
百合不是茶
泛型的继承和通配符变量的值域Object类转换
java的作用域在编程的时候经常会遇到,而我经常会搞不清楚这个
问题,所以在家的这几天回忆一下过去不知道的每个小知识点
变量的值域;
package 基础;
/**
* 作用域的范围
*
* @author Administrator
*
*/
public class zuoyongyu {
public static vo
- JDK1.5 Condition接口
bijian1013
javathreadConditionjava多线程
Condition 将 Object 监视器方法(wait、notify和 notifyAll)分解成截然不同的对象,以便通过将这些对象与任意 Lock 实现组合使用,为每个对象提供多个等待 set (wait-set)。其中,Lock 替代了 synchronized 方法和语句的使用,Condition 替代了 Object 监视器方法的使用。
条件(也称为条件队列或条件变量)为线程提供了一
- 开源中国OSC源创会记录
bijian1013
hadoopsparkMemSQL
一.Strata+Hadoop World(SHW)大会
是全世界最大的大数据大会之一。SHW大会为各种技术提供了深度交流的机会,还会看到最领先的大数据技术、最广泛的应用场景、最有趣的用例教学以及最全面的大数据行业和趋势探讨。
二.Hadoop
&nbs
- 【Java范型七】范型消除
bit1129
java
范型是Java1.5引入的语言特性,它是编译时的一个语法现象,也就是说,对于一个类,不管是范型类还是非范型类,编译得到的字节码是一样的,差别仅在于通过范型这种语法来进行编译时的类型检查,在运行时是没有范型或者类型参数这个说法的。
范型跟反射刚好相反,反射是一种运行时行为,所以编译时不能访问的变量或者方法(比如private),在运行时通过反射是可以访问的,也就是说,可见性也是一种编译时的行为,在
- 【Spark九十四】spark-sql工具的使用
bit1129
spark
spark-sql是Spark bin目录下的一个可执行脚本,它的目的是通过这个脚本执行Hive的命令,即原来通过
hive>输入的指令可以通过spark-sql>输入的指令来完成。
spark-sql可以使用内置的Hive metadata-store,也可以使用已经独立安装的Hive的metadata store
关于Hive build into Spark
- js做的各种倒计时
ronin47
js 倒计时
第一种:精确到秒的javascript倒计时代码
HTML代码:
<form name="form1">
<div align="center" align="middle"
- java-37.有n 个长为m+1 的字符串,如果某个字符串的最后m 个字符与某个字符串的前m 个字符匹配,则两个字符串可以联接
bylijinnan
java
public class MaxCatenate {
/*
* Q.37 有n 个长为m+1 的字符串,如果某个字符串的最后m 个字符与某个字符串的前m 个字符匹配,则两个字符串可以联接,
* 问这n 个字符串最多可以连成一个多长的字符串,如果出现循环,则返回错误。
*/
public static void main(String[] args){
- mongoDB安装
开窍的石头
mongodb安装 基本操作
mongoDB的安装
1:mongoDB下载 https://www.mongodb.org/downloads
2:下载mongoDB下载后解压
- [开源项目]引擎的关键意义
comsci
开源项目
一个系统,最核心的东西就是引擎。。。。。
而要设计和制造出引擎,最关键的是要坚持。。。。。。
现在最先进的引擎技术,也是从莱特兄弟那里出现的,但是中间一直没有断过研发的
- 软件度量的一些方法
cuiyadll
方法
软件度量的一些方法http://cuiyingfeng.blog.51cto.com/43841/6775/在前面我们已介绍了组成软件度量的几个方面。在这里我们将先给出关于这几个方面的一个纲要介绍。在后面我们还会作进一步具体的阐述。当我们不从高层次的概念级来看软件度量及其目标的时候,我们很容易把这些活动看成是不同而且毫不相干的。我们现在希望表明他们是怎样恰如其分地嵌入我们的框架的。也就是我们度量的
- XSD中的targetNameSpace解释
darrenzhu
xmlnamespacexsdtargetnamespace
参考链接:
http://blog.csdn.net/colin1014/article/details/357694
xsd文件中定义了一个targetNameSpace后,其内部定义的元素,属性,类型等都属于该targetNameSpace,其自身或外部xsd文件使用这些元素,属性等都必须从定义的targetNameSpace中找:
例如:以下xsd文件,就出现了该错误,即便是在一
- 什么是RAID0、RAID1、RAID0+1、RAID5,等磁盘阵列模式?
dcj3sjt126com
raid
RAID 1又称为Mirror或Mirroring,它的宗旨是最大限度的保证用户数据的可用性和可修复性。 RAID 1的操作方式是把用户写入硬盘的数据百分之百地自动复制到另外一个硬盘上。由于对存储的数据进行百分之百的备份,在所有RAID级别中,RAID 1提供最高的数据安全保障。同样,由于数据的百分之百备份,备份数据占了总存储空间的一半,因而,Mirror的磁盘空间利用率低,存储成本高。
Mir
- yii2 restful web服务快速入门
dcj3sjt126com
PHPyii2
快速入门
Yii 提供了一整套用来简化实现 RESTful 风格的 Web Service 服务的 API。 特别是,Yii 支持以下关于 RESTful 风格的 API:
支持 Active Record 类的通用API的快速原型
涉及的响应格式(在默认情况下支持 JSON 和 XML)
支持可选输出字段的定制对象序列化
适当的格式的数据采集和验证错误
- MongoDB查询(3)——内嵌文档查询(七)
eksliang
MongoDB查询内嵌文档MongoDB查询内嵌数组
MongoDB查询内嵌文档
转载请出自出处:http://eksliang.iteye.com/blog/2177301 一、概述
有两种方法可以查询内嵌文档:查询整个文档;针对键值对进行查询。这两种方式是不同的,下面我通过例子进行分别说明。
二、查询整个文档
例如:有如下文档
db.emp.insert({
&qu
- android4.4从系统图库无法加载图片的问题
gundumw100
android
典型的使用场景就是要设置一个头像,头像需要从系统图库或者拍照获得,在android4.4之前,我用的代码没问题,但是今天使用android4.4的时候突然发现不灵了。baidu了一圈,终于解决了。
下面是解决方案:
private String[] items = new String[] { "图库","拍照" };
/* 头像名称 */
- 网页特效大全 jQuery等
ini
JavaScriptjquerycsshtml5ini
HTML5和CSS3知识和特效
asp.net ajax jquery实例
分享一个下雪的特效
jQuery倾斜的动画导航菜单
选美大赛示例 你会选谁
jQuery实现HTML5时钟
功能强大的滚动播放插件JQ-Slide
万圣节快乐!!!
向上弹出菜单jQuery插件
htm5视差动画
jquery将列表倒转顺序
推荐一个jQuery分页插件
jquery animate
- swift objc_setAssociatedObject block(version1.2 xcode6.4)
啸笑天
version
import UIKit
class LSObjectWrapper: NSObject {
let value: ((barButton: UIButton?) -> Void)?
init(value: (barButton: UIButton?) -> Void) {
self.value = value
- Aegis 默认的 Xfire 绑定方式,将 XML 映射为 POJO
MagicMa_007
javaPOJOxmlAegisxfire
Aegis 是一个默认的 Xfire 绑定方式,它将 XML 映射为 POJO, 支持代码先行的开发.你开发服 务类与 POJO,它为你生成 XML schema/wsdl
XML 和 注解映射概览
默认情况下,你的 POJO 类被是基于他们的名字与命名空间被序列化。如果
- js get max value in (json) Array
qiaolevip
每天进步一点点学习永无止境max纵观千象
// Max value in Array
var arr = [1,2,3,5,3,2];Math.max.apply(null, arr); // 5
// Max value in Jaon Array
var arr = [{"x":"8/11/2009","y":0.026572007},{"x"
- XMLhttpRequest 请求 XML,JSON ,POJO 数据
Luob.
POJOjsonAjaxxmlXMLhttpREquest
在使用XMlhttpRequest对象发送请求和响应之前,必须首先使用javaScript对象创建一个XMLHttpRquest对象。
var xmlhttp;
function getXMLHttpRequest(){
if(window.ActiveXObject){
xmlhttp:new ActiveXObject("Microsoft.XMLHTTP
- jquery
wuai
jquery
以下防止文档在完全加载之前运行Jquery代码,否则会出现试图隐藏一个不存在的元素、获得未完全加载的图像的大小 等等
$(document).ready(function(){
jquery代码;
});
<script type="text/javascript" src="c:/scripts/jquery-1.4.2.min.js&quo